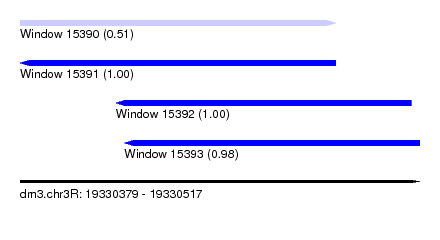
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,330,379 – 19,330,517 |
| Length | 138 |
| Max. P | 0.997405 |

| Location | 19,330,379 – 19,330,488 |
|---|---|
| Length | 109 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.68 |
| Shannon entropy | 0.42051 |
| G+C content | 0.64424 |
| Mean single sequence MFE | -36.56 |
| Consensus MFE | -13.95 |
| Energy contribution | -13.76 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.514391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
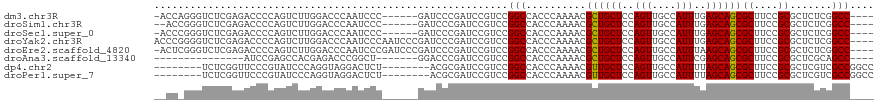
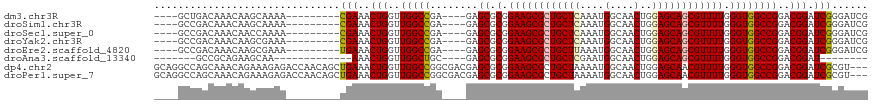
>dm3.chr3R 19330379 109 + 27905053 -ACCAGGGUCUCGAGACCCCAGUCUUGGACCCAAUCCC------GAUCCCGAUCCGUCCGGCCACCCAAAACGCUGCUCCAGUUGCCAUUUGAGCAGCGCUUCCGCGCUCUCGGCC---- -....(((((.((((((....)))))))))))......------(((....))).....((((.........(((((((.(((....))).)))))))((....))......))))---- ( -39.40, z-score = -3.03, R) >droSim1.chr3R 19150028 108 + 27517382 --ACCGGGUCUCGAGACCCCAGUCUUGGACCCAAUCCC------GAUCCCGAUCCGUCCGGCCACCCAAAACGCUGCUCCAGUUGCCAUUUGAGCAGCGCUUCCGCGCUCUCGGCC---- --...(((((.((((((....)))))))))))......------(((....))).....((((.........(((((((.(((....))).)))))))((....))......))))---- ( -39.50, z-score = -3.03, R) >droSec1.super_0 19693348 109 + 21120651 -ACCCGGGUCUCGAGACCCCAGUCUUGGACCCAAUCCC------GAUCCCGAUCCGUCCGGCCACCCAAAACGCUGCUCCAGUUGCCAUUUGAGCAGCGCUUCCGCGCUCUCGGCC---- -....(((((.((((((....)))))))))))......------(((....))).....((((.........(((((((.(((....))).)))))))((....))......))))---- ( -39.50, z-score = -3.08, R) >droYak2.chr3R 20215029 116 + 28832112 ACCCGGGGUCUCGAGACCCCAGUCUUGGACCCAAUCCCAAUCCCGAUCCCGAUCCGUCCGGCCACCCAAAACGCUGCUCCAGUUGCCAUUUGAGCAGCGCUUCCGCGCUCUCGGCC---- ...(((((((.((((((....)))))))))))............(((....)))))...((((.........(((((((.(((....))).)))))))((....))......))))---- ( -39.50, z-score = -2.40, R) >droEre2.scaffold_4820 1753013 115 - 10470090 -ACUCGGGUCUCGAGACCCCAGUCUUGGACCCAAUCCCGAUCCCGAUCCCGAUCCGUCCGGCCACCCAAAACGCUGCUCCAGUUGCCAUUUAAGCAGCGCUUCCGCGCUCUCGGCC---- -....(((((.((((((....)))))))))))......((((........)))).....((((.........((((((..(((....)))..))))))((....))......))))---- ( -38.00, z-score = -2.59, R) >droAna3.scaffold_13340 3477253 94 - 23697760 ---------------AUCCGAGCCACGAGACCCGGCU-------GGACCCGAUCCGUCCGGCCACCCAAAACGCUGCUCCAGUUGCCAUUCGAGCAGCGCUUCCGCGCUCGCAGCC---- ---------------...(((((..((.((...((((-------((((.......))))))))........((((((((.(((....))).))))))))..)))).))))).....---- ( -38.00, z-score = -3.25, R) >dp4.chr2 2818518 104 + 30794189 --------UCUCGGUUCCCGUAUCCCAGGUAGGACUCU--------ACGCGAUCCGUCCGGCCACCCAAAACGUUGCUCCAGUUGCCAUUUUAGCAGCGCUUCCGCGCUCGUCGCCGGCC --------...(((.((.(((((((......)))...)--------))).)).))).(((((.((.........((((..(((....)))..))))((((....))))..)).))))).. ( -29.30, z-score = -0.25, R) >droPer1.super_7 2911923 104 - 4445127 --------UCUCGGUUCCCGUAUCCCAGGUAGGACUCU--------ACGCGAUCCGUCCGGCCACCCAAAACGUUGCUCCAGUUGCCAUUUUAGCAGCGCUUCCGCGCUCGUCGCCGGCC --------...(((.((.(((((((......)))...)--------))).)).))).(((((.((.........((((..(((....)))..))))((((....))))..)).))))).. ( -29.30, z-score = -0.25, R) >consensus __CC_GGGUCUCGAGACCCCAGUCUUGGACCCAAUCCC______GAUCCCGAUCCGUCCGGCCACCCAAAACGCUGCUCCAGUUGCCAUUUGAGCAGCGCUUCCGCGCUCUCGGCC____ ...........................................................(((..........((((((..(((....)))..))))))((....)).......))).... (-13.95 = -13.76 + -0.19)


| Location | 19,330,379 – 19,330,488 |
|---|---|
| Length | 109 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.68 |
| Shannon entropy | 0.42051 |
| G+C content | 0.64424 |
| Mean single sequence MFE | -51.80 |
| Consensus MFE | -28.51 |
| Energy contribution | -28.62 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.997405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19330379 109 - 27905053 ----GGCCGAGAGCGCGGAAGCGCUGCUCAAAUGGCAACUGGAGCAGCGUUUUGGGUGGCCGGACGGAUCGGGAUC------GGGAUUGGGUCCAAGACUGGGGUCUCGAGACCCUGGU- ----((((((((((((....))(((((((....(....)..))))))))))))...)))))((((.((((......------..)))).).)))...((..(((((....)))))..))- ( -53.40, z-score = -3.28, R) >droSim1.chr3R 19150028 108 - 27517382 ----GGCCGAGAGCGCGGAAGCGCUGCUCAAAUGGCAACUGGAGCAGCGUUUUGGGUGGCCGGACGGAUCGGGAUC------GGGAUUGGGUCCAAGACUGGGGUCUCGAGACCCGGU-- ----.(((.(((((((....))(((((((....(....)..)))))))))))).))).(((((..(..((((((((------((..(((....)))..))..))))))))..))))))-- ( -54.00, z-score = -3.53, R) >droSec1.super_0 19693348 109 - 21120651 ----GGCCGAGAGCGCGGAAGCGCUGCUCAAAUGGCAACUGGAGCAGCGUUUUGGGUGGCCGGACGGAUCGGGAUC------GGGAUUGGGUCCAAGACUGGGGUCUCGAGACCCGGGU- ----.(((.(((((((....))(((((((....(....)..)))))))))))).)))..((((..(..((((((((------((..(((....)))..))..))))))))..)))))..- ( -53.10, z-score = -3.21, R) >droYak2.chr3R 20215029 116 - 28832112 ----GGCCGAGAGCGCGGAAGCGCUGCUCAAAUGGCAACUGGAGCAGCGUUUUGGGUGGCCGGACGGAUCGGGAUCGGGAUUGGGAUUGGGUCCAAGACUGGGGUCUCGAGACCCCGGGU ----((((((((((((....))(((((((....(....)..))))))))))))...)))))((((.((((..(((....)))..)))).).)))....((((((((....)))))))).. ( -56.60, z-score = -3.32, R) >droEre2.scaffold_4820 1753013 115 + 10470090 ----GGCCGAGAGCGCGGAAGCGCUGCUUAAAUGGCAACUGGAGCAGCGUUUUGGGUGGCCGGACGGAUCGGGAUCGGGAUCGGGAUUGGGUCCAAGACUGGGGUCUCGAGACCCGAGU- ----((((((((((((....))(((((((....(....)..))))))))))))...))))).......(((((.((((((((((..(((....)))..))..))))))))..)))))..- ( -54.20, z-score = -3.29, R) >droAna3.scaffold_13340 3477253 94 + 23697760 ----GGCUGCGAGCGCGGAAGCGCUGCUCGAAUGGCAACUGGAGCAGCGUUUUGGGUGGCCGGACGGAUCGGGUCC-------AGCCGGGUCUCGUGGCUCGGAU--------------- ----(((..(((((.(.((((((((((((.(..(....)).))))))))))))((.(((((.((....)).)).))-------).))).).))))..))).....--------------- ( -48.50, z-score = -2.48, R) >dp4.chr2 2818518 104 - 30794189 GGCCGGCGACGAGCGCGGAAGCGCUGCUAAAAUGGCAACUGGAGCAACGUUUUGGGUGGCCGGACGGAUCGCGU--------AGAGUCCUACCUGGGAUACGGGAACCGAGA-------- ..(((((.((.(((((....))))).((((((((((.......))..)))))))))).))))).(((.((.(((--------(...(((.....))).)))).)).)))...-------- ( -47.30, z-score = -3.23, R) >droPer1.super_7 2911923 104 + 4445127 GGCCGGCGACGAGCGCGGAAGCGCUGCUAAAAUGGCAACUGGAGCAACGUUUUGGGUGGCCGGACGGAUCGCGU--------AGAGUCCUACCUGGGAUACGGGAACCGAGA-------- ..(((((.((.(((((....))))).((((((((((.......))..)))))))))).))))).(((.((.(((--------(...(((.....))).)))).)).)))...-------- ( -47.30, z-score = -3.23, R) >consensus ____GGCCGAGAGCGCGGAAGCGCUGCUCAAAUGGCAACUGGAGCAGCGUUUUGGGUGGCCGGACGGAUCGGGAUC______GGGAUUGGGUCCAAGACUGGGGUCUCGAGACCC_GG__ ....((((....((.(.((((((((((((....(....)..)))))))))))).)))))))............................(..((.......))..).............. (-28.51 = -28.62 + 0.11)

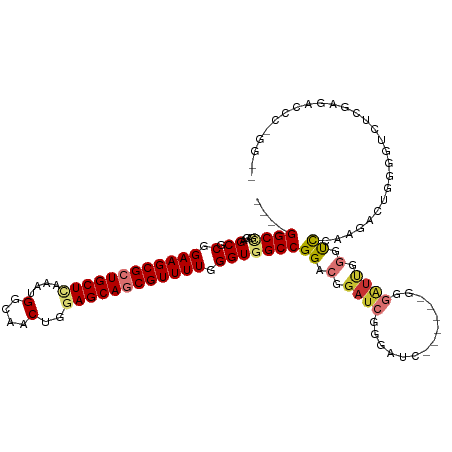
| Location | 19,330,412 – 19,330,514 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.32 |
| Shannon entropy | 0.35430 |
| G+C content | 0.59177 |
| Mean single sequence MFE | -41.44 |
| Consensus MFE | -33.29 |
| Energy contribution | -33.48 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.01 |
| SVM RNA-class probability | 0.996946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19330412 102 - 27905053 --------------GACAAACAAGCAAAACGAAACUGGUUGGCCGA----GAGCGCGGAAGCGCUGCUCAAAUGGCAACUGGAGCAGCGUUUUGGGUGGCCGGACGGAUCGGGAUCGGGA --------------...............(((..((((((.((((.----..((.(.((((((((((((....(....)..)))))))))))).)))...))).).))))))..)))... ( -43.20, z-score = -3.81, R) >droSim1.chr3R 19150060 102 - 27517382 --------------GACAAACAAGCAAAACGAAACUGGUUGGCCGA----GAGCGCGGAAGCGCUGCUCAAAUGGCAACUGGAGCAGCGUUUUGGGUGGCCGGACGGAUCGGGAUCGGGA --------------...............(((..((((((.((((.----..((.(.((((((((((((....(....)..)))))))))))).)))...))).).))))))..)))... ( -43.20, z-score = -3.81, R) >droSec1.super_0 19693381 102 - 21120651 --------------GACAAACAACCAAAACGAAACUGGUUGGCCGA----GAGCGCGGAAGCGCUGCUCAAAUGGCAACUGGAGCAGCGUUUUGGGUGGCCGGACGGAUCGGGAUCGGGA --------------...............(((..((((((.((((.----..((.(.((((((((((((....(....)..)))))))))))).)))...))).).))))))..)))... ( -43.20, z-score = -3.63, R) >droYak2.chr3R 20215069 102 - 28832112 --------------GACAAACAAGCGAAACGAAACUGGUUGGCCGA----GAGCGCGGAAGCGCUGCUCAAAUGGCAACUGGAGCAGCGUUUUGGGUGGCCGGACGGAUCGGGAUCGGGA --------------...............(((..((((((.((((.----..((.(.((((((((((((....(....)..)))))))))))).)))...))).).))))))..)))... ( -43.20, z-score = -3.57, R) >droEre2.scaffold_4820 1753052 102 + 10470090 --------------GACAAACAAGCGAAAUGAAACUGGUUGGCCGA----GAGCGCGGAAGCGCUGCUUAAAUGGCAACUGGAGCAGCGUUUUGGGUGGCCGGACGGAUCGGGAUCGGGA --------------...............(((..((((((.((((.----..((.(.((((((((((((....(....)..)))))))))))).)))...))).).))))))..)))... ( -39.20, z-score = -2.95, R) >droAna3.scaffold_13340 3477282 84 + 23697760 -----------------GCAGAAGCAA----AAACUGGUUGGCUGC----GAGCGCGGAAGCGCUGCUCGAAUGGCAACUGGAGCAGCGUUUUGGGUGGCCGGACGGAU----------- -----------------((....))..----...(((..((((..(----((((((....))(((((((.(..(....)).)))))))))))...)..))))..)))..----------- ( -35.70, z-score = -2.26, R) >dp4.chr2 2818548 113 - 30794189 GGCCAGCAAACAGAAAGAGACCAACAG-CUGAAACUGGUUGGCCGGCGACGAGCGCGGAAGCGCUGCUAAAAUGGCAACUGGAGCAACGUUUUGGGUGGCCGGACGGAUCGCGU------ .((((((...................)-))).....((((.((((((.((.(((((....))))).((((((((((.......))..)))))))))).))))).).))))))..------ ( -41.91, z-score = -1.35, R) >droPer1.super_7 2911953 113 + 4445127 GGCCAGCAAACAGAAAGAGACCAACAG-CUGAAACUGGUUGGCCGGCGACGAGCGCGGAAGCGCUGCUAAAAUGGCAACUGGAGCAACGUUUUGGGUGGCCGGACGGAUCGCGU------ .((((((...................)-))).....((((.((((((.((.(((((....))))).((((((((((.......))..)))))))))).))))).).))))))..------ ( -41.91, z-score = -1.35, R) >consensus ______________GACAAACAAGCAAAACGAAACUGGUUGGCCGA____GAGCGCGGAAGCGCUGCUCAAAUGGCAACUGGAGCAGCGUUUUGGGUGGCCGGACGGAUCGGGAUCGGGA .............................(((..(((..((((((.....((((((....))(((((((....(....)..)))))))))))....))))))..))).)))......... (-33.29 = -33.48 + 0.19)



| Location | 19,330,415 – 19,330,517 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.88 |
| Shannon entropy | 0.33281 |
| G+C content | 0.59884 |
| Mean single sequence MFE | -41.55 |
| Consensus MFE | -33.20 |
| Energy contribution | -33.51 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.980990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19330415 102 - 27905053 ----GCUGACAAACAAGCAAAA---------CGAAACUGGUUGGCCGA----GAGCGCGGAAGCGCUGCUCAAAUGGCAACUGGAGCAGCGUUUUGGGUGGCCGGACGGAUCGGGAUCG ----(((........)))....---------(((..((((((.((((.----..((.(.((((((((((((....(....)..)))))))))))).)))...))).).))))))..))) ( -44.90, z-score = -3.95, R) >droSim1.chr3R 19150063 102 - 27517382 ----GCCGACAAACAAGCAAAA---------CGAAACUGGUUGGCCGA----GAGCGCGGAAGCGCUGCUCAAAUGGCAACUGGAGCAGCGUUUUGGGUGGCCGGACGGAUCGGGAUCG ----..................---------(((..((((((.((((.----..((.(.((((((((((((....(....)..)))))))))))).)))...))).).))))))..))) ( -41.90, z-score = -2.63, R) >droSec1.super_0 19693384 102 - 21120651 ----GCCGACAAACAACCAAAA---------CGAAACUGGUUGGCCGA----GAGCGCGGAAGCGCUGCUCAAAUGGCAACUGGAGCAGCGUUUUGGGUGGCCGGACGGAUCGGGAUCG ----.((((....((((((...---------......))))))(((.(----((((((....))(((((((....(....)..)))))))))))).)))..((....)).))))..... ( -42.80, z-score = -2.93, R) >droYak2.chr3R 20215072 102 - 28832112 ----GCCGACAAACAAGCGAAA---------CGAAACUGGUUGGCCGA----GAGCGCGGAAGCGCUGCUCAAAUGGCAACUGGAGCAGCGUUUUGGGUGGCCGGACGGAUCGGGAUCG ----..................---------(((..((((((.((((.----..((.(.((((((((((((....(....)..)))))))))))).)))...))).).))))))..))) ( -41.90, z-score = -2.33, R) >droEre2.scaffold_4820 1753055 102 + 10470090 ----GCCGACAAACAAGCGAAA---------UGAAACUGGUUGGCCGA----GAGCGCGGAAGCGCUGCUUAAAUGGCAACUGGAGCAGCGUUUUGGGUGGCCGGACGGAUCGGGAUCG ----.((((....((.......---------))...((((.(.(((.(----((((((....))(((((((....(....)..)))))))))))).))).))))).....))))..... ( -38.90, z-score = -2.04, R) >droAna3.scaffold_13340 3477282 87 + 23697760 -------GCCGCAGAAGCAA-------------AAACUGGUUGGCUGC----GAGCGCGGAAGCGCUGCUCGAAUGGCAACUGGAGCAGCGUUUUGGGUGGCCGGACGGAU-------- -------(((((((.(((..-------------......)))..))))----).)).(.((((((((((((.(..(....)).)))))))))))).)....((....))..-------- ( -36.00, z-score = -1.24, R) >dp4.chr2 2818548 116 - 30794189 GCAGGCCAGCAAACAGAAAGAGACCAACAGCUGAAACUGGUUGGCCGGCGACGAGCGCGGAAGCGCUGCUAAAAUGGCAACUGGAGCAACGUUUUGGGUGGCCGGACGGAUCGCGU--- ((.(((((((.........(....)..(((......))))))))))(((.((.(((((....))))).((((((((((.......))..)))))))))).))).........))..--- ( -43.00, z-score = -1.22, R) >droPer1.super_7 2911953 116 + 4445127 GCAGGCCAGCAAACAGAAAGAGACCAACAGCUGAAACUGGUUGGCCGGCGACGAGCGCGGAAGCGCUGCUAAAAUGGCAACUGGAGCAACGUUUUGGGUGGCCGGACGGAUCGCGU--- ((.(((((((.........(....)..(((......))))))))))(((.((.(((((....))))).((((((((((.......))..)))))))))).))).........))..--- ( -43.00, z-score = -1.22, R) >consensus ____GCCGACAAACAAGCAAAA_________CGAAACUGGUUGGCCGA____GAGCGCGGAAGCGCUGCUCAAAUGGCAACUGGAGCAGCGUUUUGGGUGGCCGGACGGAUCGGGAUCG ....................................(((..((((((.....((((((....))(((((((....(....)..)))))))))))....))))))..))).......... (-33.20 = -33.51 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:38:20 2011