| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,322,135 – 19,322,245 |
| Length | 110 |
| Max. P | 0.999719 |

| Location | 19,322,135 – 19,322,237 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 84.97 |
| Shannon entropy | 0.24536 |
| G+C content | 0.47370 |
| Mean single sequence MFE | -41.00 |
| Consensus MFE | -34.34 |
| Energy contribution | -35.65 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.97 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.59 |
| SVM RNA-class probability | 0.999004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
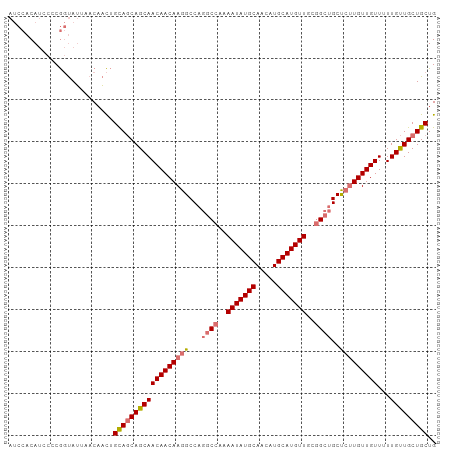
>dm3.chr3R 19322135 102 + 27905053 AUCCACAUACCCGGUAUUAACAACUGCAGCAGCAACAACAAGGCCAGGCCGAAAUAUGCAACAUGCAUGUUGCGGCUGCUCUUGUUGUUUUUGUUGCUGCUG .........................(((((((((((((((((((..(((((.(((((((.....))))))).))))))).))))))))...))))))))).. ( -43.60, z-score = -4.59, R) >droSim1.chr3R 19141415 102 + 27517382 AUCCACAUCCCCGGUAUUAACAACUGCAGCAGCAACAACAAGGCCAGGCCAAAAUAUGCAACAUGCAUGUUGCGGCUGCUCUUGUUGUUUUUGUUGCUGCUG .........................(((((((((((((((((((..((((..(((((((.....)))))))..)))))).))))))))...))))))))).. ( -41.00, z-score = -3.90, R) >droSec1.super_0 19685127 102 + 21120651 AUCCACAUGCCCGGUAUUAACAACUGCAGCAGCAACAACAAGGCCAGCCCAAAAUAUGCAACAUGCAUGUUGCGGCUGCUCUUGUUGUUUUUGUUGCUGCUG .........................(((((((((((((((((..(((((...(((((((.....)))))))..)))))..))))))))...))))))))).. ( -43.40, z-score = -4.41, R) >droAna3.scaffold_13340 3468620 97 - 23697760 -----CAACAACACCAGUAACAUUGGUAACAGCAACAACAAGGCCAAGCCAAAAUAUGCAACAUGCAUGUUAUUGCGGCCGAUGUUGUUGUUGCUGCUGCUG -----.........((((......((((.(((((((((((.((((..((...(((((((.....)))))))...))))))..))))))))))).)))))))) ( -36.00, z-score = -3.00, R) >consensus AUCCACAUCCCCGGUAUUAACAACUGCAGCAGCAACAACAAGGCCAGGCCAAAAUAUGCAACAUGCAUGUUGCGGCUGCUCUUGUUGUUUUUGUUGCUGCUG .........................((((((((((((((((((...(((((.(((((((.....))))))).)))))..)))))))))...))))))))).. (-34.34 = -35.65 + 1.31)


| Location | 19,322,135 – 19,322,237 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 84.97 |
| Shannon entropy | 0.24536 |
| G+C content | 0.47370 |
| Mean single sequence MFE | -37.05 |
| Consensus MFE | -29.00 |
| Energy contribution | -29.50 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.737565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19322135 102 - 27905053 CAGCAGCAACAAAAACAACAAGAGCAGCCGCAACAUGCAUGUUGCAUAUUUCGGCCUGGCCUUGUUGUUGCUGCUGCAGUUGUUAAUACCGGGUAUGUGGAU (((((((((((......(((((.((.((((.((.((((.....)))).)).))))...))))))))))))))))))............(((......))).. ( -37.30, z-score = -1.69, R) >droSim1.chr3R 19141415 102 - 27517382 CAGCAGCAACAAAAACAACAAGAGCAGCCGCAACAUGCAUGUUGCAUAUUUUGGCCUGGCCUUGUUGUUGCUGCUGCAGUUGUUAAUACCGGGGAUGUGGAU (((((((((((......(((((.((.((((.((.((((.....)))).)).))))...))))))))))))))))))............(((......))).. ( -35.70, z-score = -1.05, R) >droSec1.super_0 19685127 102 - 21120651 CAGCAGCAACAAAAACAACAAGAGCAGCCGCAACAUGCAUGUUGCAUAUUUUGGGCUGGCCUUGUUGUUGCUGCUGCAGUUGUUAAUACCGGGCAUGUGGAU (((((((((((......(((((..(((((.(((.((((.....))))...))))))))..))))))))))))))))((..((((.......))))..))... ( -37.50, z-score = -1.24, R) >droAna3.scaffold_13340 3468620 97 + 23697760 CAGCAGCAGCAACAACAACAUCGGCCGCAAUAACAUGCAUGUUGCAUAUUUUGGCUUGGCCUUGUUGUUGCUGUUACCAAUGUUACUGGUGUUGUUG----- ((((((((((((((((((....((((((...((.((((.....)))).))...))..))))))))))))))))))((((.......))))))))...----- ( -37.70, z-score = -2.80, R) >consensus CAGCAGCAACAAAAACAACAAGAGCAGCCGCAACAUGCAUGUUGCAUAUUUUGGCCUGGCCUUGUUGUUGCUGCUGCAGUUGUUAAUACCGGGGAUGUGGAU (((((((((((......(((((.((.((((.((.((((.....)))).)).))))...)))))))))))))))))).......................... (-29.00 = -29.50 + 0.50)


| Location | 19,322,143 – 19,322,245 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 72.78 |
| Shannon entropy | 0.52550 |
| G+C content | 0.48481 |
| Mean single sequence MFE | -40.61 |
| Consensus MFE | -22.23 |
| Energy contribution | -24.13 |
| Covariance contribution | 1.90 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.53 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.25 |
| SVM RNA-class probability | 0.999719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
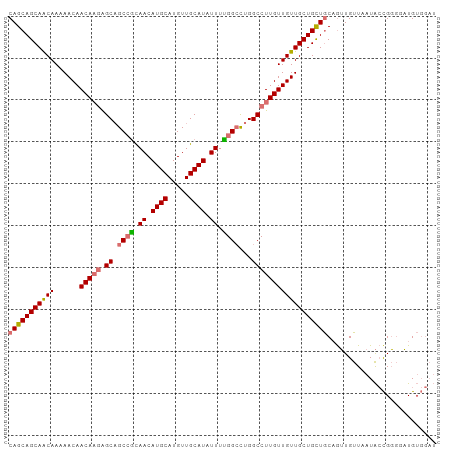
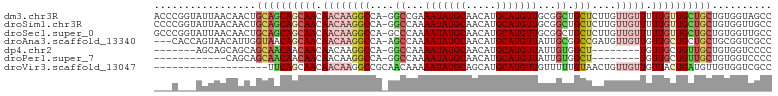
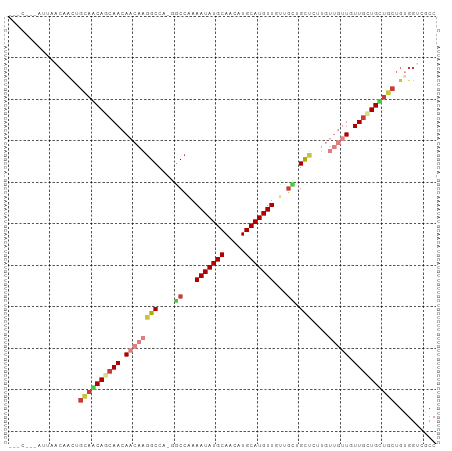
>dm3.chr3R 19322143 102 + 27905053 ACCCGGUAUUAACAACUGCAGCAGCAACAACAAGGCCA-GGCCGAAAUAUGCAACAUGCAUGUUGCGGCUGCUCUUGUUGUUUUUGUUGCUGCUGUGGUAGCC ..............((..((((((((((((((((((..-(((((.(((((((.....))))))).))))))).)))))......)))))))))))..)).... ( -47.30, z-score = -4.32, R) >droSim1.chr3R 19141423 102 + 27517382 CCCCGGUAUUAACAACUGCAGCAGCAACAACAAGGCCA-GGCCAAAAUAUGCAACAUGCAUGUUGCGGCUGCUCUUGUUGUUUUUGUUGCUGCUGUGGUUGCC ............((((..((((((((((((((((((..-((((..(((((((.....)))))))..)))))).)))))......)))))))))))..)))).. ( -48.10, z-score = -4.50, R) >droSec1.super_0 19685135 102 + 21120651 GCCCGGUAUUAACAACUGCAGCAGCAACAACAAGGCCA-GCCCAAAAUAUGCAACAUGCAUGUUGCGGCUGCUCUUGUUGUUUUUGUUGCUGCUGUGGUUGCC ............((((..((((((((((((((((..((-(((...(((((((.....)))))))..)))))..)))))......)))))))))))..)))).. ( -50.50, z-score = -5.27, R) >droAna3.scaffold_13340 3468626 99 - 23697760 ---CACCAGUAACAUUGGUAACAGCAACAACAAGGCCA-AGCCAAAAUAUGCAACAUGCAUGUUAUUGCGGCCGAUGUUGUUGUUGCUGCUGCUGCGGUCGCC ---.(((.(((.((..(((((((((((((....((((.-.((...(((((((.....)))))))...))))))..)))))))))))))..)).)))))).... ( -39.20, z-score = -2.68, R) >dp4.chr2 2808998 87 + 30794189 -------AGCAGCAGCAGCAACAACAACAACAAGGCCA-GGCCAAAAUAUGCAACAUGCAUGUUAUUGUGGCU--------UGUUGCUGUUGCUGUGGUCCCC -------.((((((((((((((((..((((...(((..-.)))..(((((((.....))))))).))))...)--------)))))))))))))))....... ( -39.40, z-score = -3.60, R) >droPer1.super_7 2902344 82 - 4445127 ------------CAGCAGCAACAACAACAACAAGGCCA-GGCCAAAAUAUGCAACAUGCAUGUUAUUGUGGCU--------UGUUGCUGUUGCUGUGGUCCCC ------------..(((((((((.(((((....(((((-...((((((((((.....))))))).))))))))--------))))).)))))))))....... ( -32.90, z-score = -2.57, R) >droVir3.scaffold_13047 8951049 84 + 19223366 -------------------UUCAGCAACAACAAGGCCGCAACAAAAAUAUGCAGCAUGCAUGUUGUUUUUGUAACUGUUGUUGUUACUGAUGUUGUGGUCGCC -------------------..............((((((((((..(((((((.....)))))))......(((((.......)))))...))))))))))... ( -26.90, z-score = -1.79, R) >consensus ___C___AUUAACAACUGCAACAGCAACAACAAGGCCA_GGCCAAAAUAUGCAACAUGCAUGUUGUUGCUGCUCUUGUUGUUGUUGCUGCUGCUGUGGUCGCC .................((((((((((.((((((((....)))..(((((((.....))))))).............))))).)))))))))).......... (-22.23 = -24.13 + 1.90)

| Location | 19,322,143 – 19,322,245 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 72.78 |
| Shannon entropy | 0.52550 |
| G+C content | 0.48481 |
| Mean single sequence MFE | -35.89 |
| Consensus MFE | -21.59 |
| Energy contribution | -21.37 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.994250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
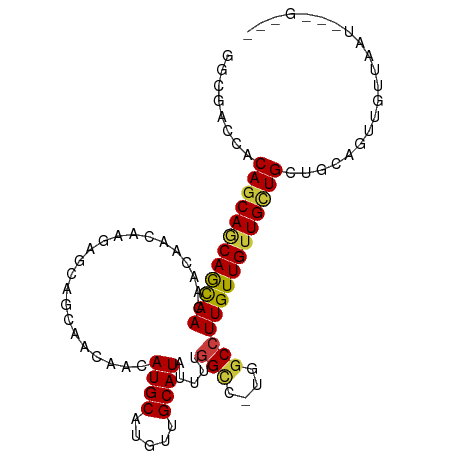
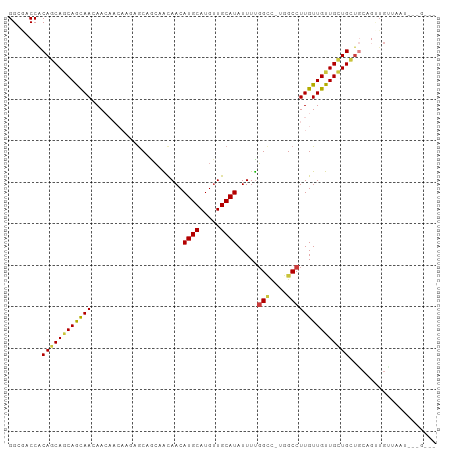
>dm3.chr3R 19322143 102 - 27905053 GGCUACCACAGCAGCAACAAAAACAACAAGAGCAGCCGCAACAUGCAUGUUGCAUAUUUCGGCC-UGGCCUUGUUGUUGCUGCUGCAGUUGUUAAUACCGGGU (((.((..(((((((((((......(((((.((.((((.((.((((.....)))).)).)))).-..))))))))))))))))))..)).))).......... ( -39.00, z-score = -2.10, R) >droSim1.chr3R 19141423 102 - 27517382 GGCAACCACAGCAGCAACAAAAACAACAAGAGCAGCCGCAACAUGCAUGUUGCAUAUUUUGGCC-UGGCCUUGUUGUUGCUGCUGCAGUUGUUAAUACCGGGG ((((((..(((((((((((......(((((.((.((((.((.((((.....)))).)).)))).-..))))))))))))))))))..)))))).......... ( -41.50, z-score = -2.64, R) >droSec1.super_0 19685135 102 - 21120651 GGCAACCACAGCAGCAACAAAAACAACAAGAGCAGCCGCAACAUGCAUGUUGCAUAUUUUGGGC-UGGCCUUGUUGUUGCUGCUGCAGUUGUUAAUACCGGGC ((((((..(((((((((((......(((((..(((((.(((.((((.....))))...))))))-))..))))))))))))))))..)))))).......... ( -43.10, z-score = -2.92, R) >droAna3.scaffold_13340 3468626 99 + 23697760 GGCGACCGCAGCAGCAGCAACAACAACAUCGGCCGCAAUAACAUGCAUGUUGCAUAUUUUGGCU-UGGCCUUGUUGUUGCUGUUACCAAUGUUACUGGUG--- .((....))...((((((((((((((....((((((...((.((((.....)))).))...)).-.))))))))))))))))))((((.......)))).--- ( -36.30, z-score = -1.76, R) >dp4.chr2 2808998 87 - 30794189 GGGGACCACAGCAACAGCAACA--------AGCCACAAUAACAUGCAUGUUGCAUAUUUUGGCC-UGGCCUUGUUGUUGUUGUUGCUGCUGCUGCU------- (....)..(((((.((((((((--------....((((((((((((.....)))).....(((.-..)))..)))))))))))))))).)))))..------- ( -33.70, z-score = -1.52, R) >droPer1.super_7 2902344 82 + 4445127 GGGGACCACAGCAACAGCAACA--------AGCCACAAUAACAUGCAUGUUGCAUAUUUUGGCC-UGGCCUUGUUGUUGUUGUUGCUGCUG------------ (....)((((((((((((((((--------((((((((....((((.....))))...)))...-))).)))))))))))))))).))...------------ ( -31.40, z-score = -1.85, R) >droVir3.scaffold_13047 8951049 84 - 19223366 GGCGACCACAACAUCAGUAACAACAACAGUUACAAAAACAACAUGCAUGCUGCAUAUUUUUGUUGCGGCCUUGUUGUUGCUGAA------------------- .............(((((((((((((..((.(((((((....((((.....)))).))))))).))....))))))))))))).------------------- ( -26.20, z-score = -2.43, R) >consensus GGCGACCACAGCAGCAGCAACAACAACAAGAGCAGCAACAACAUGCAUGUUGCAUAUUUUGGCC_UGGCCUUGUUGUUGCUGCUGCAGUUGUUAAU___G___ ........((((((((((((......................((((.....)))).....(((....)))))))))))))))..................... (-21.59 = -21.37 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:38:17 2011