
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,320,849 – 19,320,953 |
| Length | 104 |
| Max. P | 0.998484 |

| Location | 19,320,849 – 19,320,941 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 88.25 |
| Shannon entropy | 0.23875 |
| G+C content | 0.36510 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -19.45 |
| Energy contribution | -19.64 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.996455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
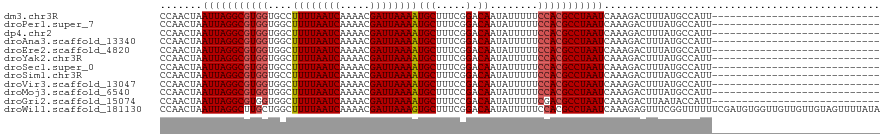
>dm3.chr3R 19320849 92 + 27905053 ---------------------GGCAGACAGUUUCCAACUAAUUAGGCGUGGUGCCUUUUAAUCAAAACGAUUAAAAUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAAGA ---------------------...................(((((((((((.((.((((((((.....)))))))).)).................)))))))))))...... ( -23.87, z-score = -3.14, R) >dp4.chr2 2807638 113 + 30794189 ACAGCUAACAGAGGACAUAUAGGCAGACAGUUUCCAACUAAUUAGGCGUGGUGGCUUUUAAUCAAAACGAUUAAAAUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAAGA ............(((...((.(.....).)).))).....(((((((((((.(((((((((((.....)))))))).)))................)))))))))))...... ( -26.03, z-score = -2.22, R) >droAna3.scaffold_13340 3467331 92 - 23697760 ---------------------GGCACACAAUUUCCAACUAAUUAGGCGUGGUGGCUUUUAAUCAAAACGAUUAAAAUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAAGA ---------------------...................(((((((((((.(((((((((((.....)))))))).)))................)))))))))))...... ( -24.73, z-score = -4.11, R) >droEre2.scaffold_4820 1743442 92 - 10470090 ---------------------UGCAGACAGUUUCCAACUAAUUAGGCGUGGUGGCUUUUAAUCAAAACGAUUAAAAUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAAGA ---------------------...................(((((((((((.(((((((((((.....)))))))).)))................)))))))))))...... ( -24.73, z-score = -3.36, R) >droYak2.chr3R 20205026 92 + 28832112 ---------------------UGCAGACAGUUUCCAACUAAUUAGGCGUGGUGGCUUUUAAUCAAAACGAUUAAAAUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAAGA ---------------------...................(((((((((((.(((((((((((.....)))))))).)))................)))))))))))...... ( -24.73, z-score = -3.36, R) >droSec1.super_0 19683908 92 + 21120651 ---------------------GGCAGACAGUUUCCAACUAAUUAGGCGUGGUGCCUUUUAAUCAAAACGAUUAAAAUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAAGA ---------------------...................(((((((((((.((.((((((((.....)))))))).)).................)))))))))))...... ( -23.87, z-score = -3.14, R) >droSim1.chr3R 19140137 92 + 27517382 ---------------------GGCAGACAGUUUCCAACUAAUUAGGCGUGGUGCCUUUUAAUCAAAACGAUUAAAAUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAAGA ---------------------...................(((((((((((.((.((((((((.....)))))))).)).................)))))))))))...... ( -23.87, z-score = -3.14, R) >droVir3.scaffold_13047 8949869 94 + 19223366 ------------------UUUUAGU-AAAAUUUCCAACUAAUUAGGCGUGGUGGCUUUUAAUCAAAACGAUUAAAAUGCUUUCCGACAAUAUUUUUCCACGCCUAAUCAAAGA ------------------..(((((-..........)))))((((((((((.(((((((((((.....)))))))).)))................))))))))))....... ( -24.93, z-score = -4.49, R) >droMoj3.scaffold_6540 2435898 99 - 34148556 --------------UGCCGAAAAGCGAAAACUUCCAACUAAUUAGGCGUGGUGGCUUUUAAUCAAAACGAUUAAAAUGCUUUCCGACAAUAUUUUUCCACGCCUAAUCAAAGA --------------.((......))...............(((((((((((.(((((((((((.....)))))))).)))................)))))))))))...... ( -25.43, z-score = -3.36, R) >droGri2.scaffold_15074 7059619 94 - 7742996 -------------------UUGCAUGAAAAUUUCCAACUAAUUAGGCGUGGUGGCUUUUAAUCAAAACGAUUAAAAUGCUUUCCGACAAUAUUUUUCGACGCCUAAUCAAAGA -------------------.....................(((((((((((.(((((((((((.....)))))))).)))..))((.........)).)))))))))...... ( -21.90, z-score = -2.60, R) >droWil1.scaffold_181130 9373784 88 + 16660200 -------------------------UCCAAUUUCCAACUAAUUAGGCUUGCUGGCUUUUAAUCAAAACGAUUAAAGUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAAGA -------------------------...............(((((((..(..(((((((((((.....)))))))).)))..)(((.........)))..)))))))...... ( -18.80, z-score = -2.75, R) >consensus _____________________GGCAGACAGUUUCCAACUAAUUAGGCGUGGUGGCUUUUAAUCAAAACGAUUAAAAUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAAGA ........................................(((((((((((....((((((((.....))))))))(((.....).))........)))))))))))...... (-19.45 = -19.64 + 0.19)


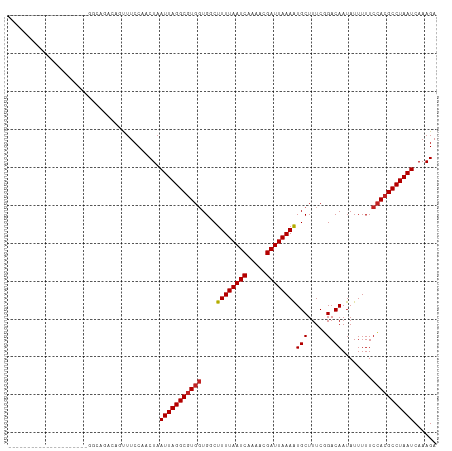
| Location | 19,320,849 – 19,320,941 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 88.25 |
| Shannon entropy | 0.23875 |
| G+C content | 0.36510 |
| Mean single sequence MFE | -27.36 |
| Consensus MFE | -25.25 |
| Energy contribution | -25.61 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.45 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.37 |
| SVM RNA-class probability | 0.998484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19320849 92 - 27905053 UCUUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCAUUUUAAUCGUUUUGAUUAAAAGGCACCACGCCUAAUUAGUUGGAAACUGUCUGCC--------------------- ...((((((((((((((.......((....))..((.((((((((.....)))))))).)).))))))))))))))..(....)........--------------------- ( -28.90, z-score = -3.99, R) >dp4.chr2 2807638 113 - 30794189 UCUUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCAUUUUAAUCGUUUUGAUUAAAAGCCACCACGCCUAAUUAGUUGGAAACUGUCUGCCUAUAUGUCCUCUGUUAGCUGU ...((((((((((((((............(....)..((((((((.....))))))))....))))))))))))))..(((...(((......)))..)))............ ( -28.40, z-score = -2.61, R) >droAna3.scaffold_13340 3467331 92 + 23697760 UCUUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCAUUUUAAUCGUUUUGAUUAAAAGCCACCACGCCUAAUUAGUUGGAAAUUGUGUGCC--------------------- (((((((((((((((((............(....)..((((((((.....))))))))....))))))))))))))..)))...........--------------------- ( -27.80, z-score = -3.98, R) >droEre2.scaffold_4820 1743442 92 + 10470090 UCUUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCAUUUUAAUCGUUUUGAUUAAAAGCCACCACGCCUAAUUAGUUGGAAACUGUCUGCA--------------------- ...((((((((((((((............(....)..((((((((.....))))))))....))))))))))))))..(....)........--------------------- ( -28.10, z-score = -3.97, R) >droYak2.chr3R 20205026 92 - 28832112 UCUUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCAUUUUAAUCGUUUUGAUUAAAAGCCACCACGCCUAAUUAGUUGGAAACUGUCUGCA--------------------- ...((((((((((((((............(....)..((((((((.....))))))))....))))))))))))))..(....)........--------------------- ( -28.10, z-score = -3.97, R) >droSec1.super_0 19683908 92 - 21120651 UCUUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCAUUUUAAUCGUUUUGAUUAAAAGGCACCACGCCUAAUUAGUUGGAAACUGUCUGCC--------------------- ...((((((((((((((.......((....))..((.((((((((.....)))))))).)).))))))))))))))..(....)........--------------------- ( -28.90, z-score = -3.99, R) >droSim1.chr3R 19140137 92 - 27517382 UCUUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCAUUUUAAUCGUUUUGAUUAAAAGGCACCACGCCUAAUUAGUUGGAAACUGUCUGCC--------------------- ...((((((((((((((.......((....))..((.((((((((.....)))))))).)).))))))))))))))..(....)........--------------------- ( -28.90, z-score = -3.99, R) >droVir3.scaffold_13047 8949869 94 - 19223366 UCUUUGAUUAGGCGUGGAAAAAUAUUGUCGGAAAGCAUUUUAAUCGUUUUGAUUAAAAGCCACCACGCCUAAUUAGUUGGAAAUUUU-ACUAAAA------------------ (((((((((((((((((........(((......)))((((((((.....))))))))....))))))))))))))..)))......-.......------------------ ( -27.20, z-score = -4.04, R) >droMoj3.scaffold_6540 2435898 99 + 34148556 UCUUUGAUUAGGCGUGGAAAAAUAUUGUCGGAAAGCAUUUUAAUCGUUUUGAUUAAAAGCCACCACGCCUAAUUAGUUGGAAGUUUUCGCUUUUCGGCA-------------- ....(((((((((((((........(((......)))((((((((.....))))))))....)))))))))))))(((((((((....)).))))))).-------------- ( -31.40, z-score = -3.62, R) >droGri2.scaffold_15074 7059619 94 + 7742996 UCUUUGAUUAGGCGUCGAAAAAUAUUGUCGGAAAGCAUUUUAAUCGUUUUGAUUAAAAGCCACCACGCCUAAUUAGUUGGAAAUUUUCAUGCAA------------------- ((((((((((((((((((......)))..((...((.((((((((.....))))))))))..))))))))))))))..))).............------------------- ( -22.80, z-score = -2.27, R) >droWil1.scaffold_181130 9373784 88 - 16660200 UCUUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCACUUUAAUCGUUUUGAUUAAAAGCCAGCAAGCCUAAUUAGUUGGAAAUUGGA------------------------- (((((((((((((.(((.........((.(....).))(((((((.....)))))))..)))....))))))))))..))).......------------------------- ( -20.50, z-score = -1.51, R) >consensus UCUUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCAUUUUAAUCGUUUUGAUUAAAAGCCACCACGCCUAAUUAGUUGGAAACUGUCUGCC_____________________ (((((((((((((((((........(((......)))((((((((.....))))))))....))))))))))))))..)))................................ (-25.25 = -25.61 + 0.36)


| Location | 19,320,861 – 19,320,953 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.26 |
| Shannon entropy | 0.15146 |
| G+C content | 0.35707 |
| Mean single sequence MFE | -24.39 |
| Consensus MFE | -19.58 |
| Energy contribution | -19.76 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.86 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.997411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
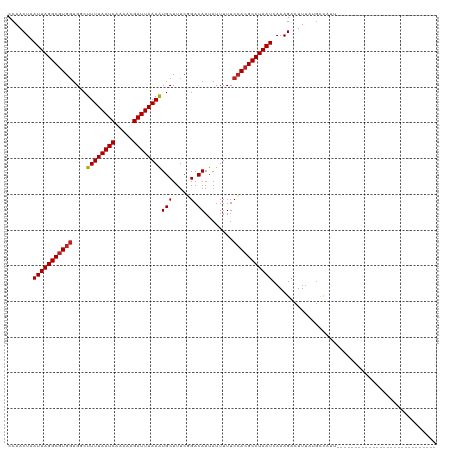
>dm3.chr3R 19320861 92 + 27905053 CCAACUAAUUAGGCGUGGUGCCUUUUAAUCAAAACGAUUAAAAUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAAGACUUUAUGCCAUU---------------------------- .......(((((((((((.((.((((((((.....)))))))).)).................)))))))))))..................---------------------------- ( -23.87, z-score = -4.14, R) >droPer1.super_7 2901024 92 - 4445127 CCAACUAAUUAGGCGUGGUGGCUUUUAAUCAAAACGAUUAAAAUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAAGACUUUAUGCCAUU---------------------------- .......(((((((((((.(((((((((((.....)))))))).)))................)))))))))))..................---------------------------- ( -24.73, z-score = -4.02, R) >dp4.chr2 2807671 92 + 30794189 CCAACUAAUUAGGCGUGGUGGCUUUUAAUCAAAACGAUUAAAAUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAAGACUUUAUGCCAUU---------------------------- .......(((((((((((.(((((((((((.....)))))))).)))................)))))))))))..................---------------------------- ( -24.73, z-score = -4.02, R) >droAna3.scaffold_13340 3467343 92 - 23697760 CCAACUAAUUAGGCGUGGUGGCUUUUAAUCAAAACGAUUAAAAUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAAGACUUUAUGCCAUU---------------------------- .......(((((((((((.(((((((((((.....)))))))).)))................)))))))))))..................---------------------------- ( -24.73, z-score = -4.02, R) >droEre2.scaffold_4820 1743454 92 - 10470090 CCAACUAAUUAGGCGUGGUGGCUUUUAAUCAAAACGAUUAAAAUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAAGACUUUAUGCCAUU---------------------------- .......(((((((((((.(((((((((((.....)))))))).)))................)))))))))))..................---------------------------- ( -24.73, z-score = -4.02, R) >droYak2.chr3R 20205038 92 + 28832112 CCAACUAAUUAGGCGUGGUGGCUUUUAAUCAAAACGAUUAAAAUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAAGACUUUAUGCCAUU---------------------------- .......(((((((((((.(((((((((((.....)))))))).)))................)))))))))))..................---------------------------- ( -24.73, z-score = -4.02, R) >droSec1.super_0 19683920 92 + 21120651 CCAACUAAUUAGGCGUGGUGCCUUUUAAUCAAAACGAUUAAAAUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAAGACUUUAUGCCAUU---------------------------- .......(((((((((((.((.((((((((.....)))))))).)).................)))))))))))..................---------------------------- ( -23.87, z-score = -4.14, R) >droSim1.chr3R 19140149 92 + 27517382 CCAACUAAUUAGGCGUGGUGCCUUUUAAUCAAAACGAUUAAAAUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAAGACUUUAUGCCAUU---------------------------- .......(((((((((((.((.((((((((.....)))))))).)).................)))))))))))..................---------------------------- ( -23.87, z-score = -4.14, R) >droVir3.scaffold_13047 8949883 92 + 19223366 CCAACUAAUUAGGCGUGGUGGCUUUUAAUCAAAACGAUUAAAAUGCUUUCCGACAAUAUUUUUCCACGCCUAAUCAAAGACUUUAUGCCAUU---------------------------- .......(((((((((((.(((((((((((.....)))))))).)))................)))))))))))..................---------------------------- ( -24.73, z-score = -4.46, R) >droMoj3.scaffold_6540 2435917 92 - 34148556 CCAACUAAUUAGGCGUGGUGGCUUUUAAUCAAAACGAUUAAAAUGCUUUCCGACAAUAUUUUUCCACGCCUAAUCAAAGACUUUAUGCCAUU---------------------------- .......(((((((((((.(((((((((((.....)))))))).)))................)))))))))))..................---------------------------- ( -24.73, z-score = -4.46, R) >droGri2.scaffold_15074 7059633 92 - 7742996 CCAACUAAUUAGGCGUGGUGGCUUUUAAUCAAAACGAUUAAAAUGCUUUCCGACAAUAUUUUUCGACGCCUAAUCAAAGACUUAAUACCAUU---------------------------- .......(((((((((((.(((((((((((.....)))))))).)))..))((.........)).)))))))))..................---------------------------- ( -21.90, z-score = -3.73, R) >droWil1.scaffold_181130 9373792 120 + 16660200 CCAACUAAUUAGGCUUGCUGGCUUUUAAUCAAAACGAUUAAAGUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAAGAGUUUCGGUUUUUUCGAUGUGGUUGUUGUUGUAGUUUUAUA .(((((((((((((..(..(((((((((((.....)))))))).)))..)(((.........)))..))))))).........((((.....))))..))))))................ ( -26.00, z-score = -1.10, R) >consensus CCAACUAAUUAGGCGUGGUGGCUUUUAAUCAAAACGAUUAAAAUGCUUUCGGACAAUAUUUUUCCACGCCUAAUCAAAGACUUUAUGCCAUU____________________________ .......(((((((((((....((((((((.....))))))))(((.....).))........))))))))))).............................................. (-19.58 = -19.76 + 0.17)

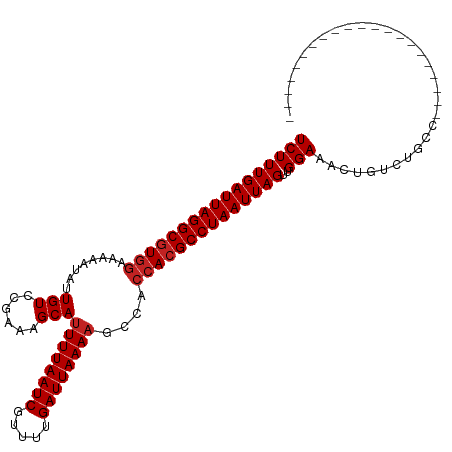
| Location | 19,320,861 – 19,320,953 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.26 |
| Shannon entropy | 0.15146 |
| G+C content | 0.35707 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -24.79 |
| Energy contribution | -25.14 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.75 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.32 |
| SVM RNA-class probability | 0.998307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19320861 92 - 27905053 ----------------------------AAUGGCAUAAAGUCUUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCAUUUUAAUCGUUUUGAUUAAAAGGCACCACGCCUAAUUAGUUGG ----------------------------...(((.....))).((((((((((((((.......((....))..((.((((((((.....)))))))).)).)))))))))))))).... ( -29.00, z-score = -4.17, R) >droPer1.super_7 2901024 92 + 4445127 ----------------------------AAUGGCAUAAAGUCUUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCAUUUUAAUCGUUUUGAUUAAAAGCCACCACGCCUAAUUAGUUGG ----------------------------...(((.....))).((((((((((((((............(....)..((((((((.....))))))))....)))))))))))))).... ( -28.20, z-score = -4.06, R) >dp4.chr2 2807671 92 - 30794189 ----------------------------AAUGGCAUAAAGUCUUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCAUUUUAAUCGUUUUGAUUAAAAGCCACCACGCCUAAUUAGUUGG ----------------------------...(((.....))).((((((((((((((............(....)..((((((((.....))))))))....)))))))))))))).... ( -28.20, z-score = -4.06, R) >droAna3.scaffold_13340 3467343 92 + 23697760 ----------------------------AAUGGCAUAAAGUCUUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCAUUUUAAUCGUUUUGAUUAAAAGCCACCACGCCUAAUUAGUUGG ----------------------------...(((.....))).((((((((((((((............(....)..((((((((.....))))))))....)))))))))))))).... ( -28.20, z-score = -4.06, R) >droEre2.scaffold_4820 1743454 92 + 10470090 ----------------------------AAUGGCAUAAAGUCUUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCAUUUUAAUCGUUUUGAUUAAAAGCCACCACGCCUAAUUAGUUGG ----------------------------...(((.....))).((((((((((((((............(....)..((((((((.....))))))))....)))))))))))))).... ( -28.20, z-score = -4.06, R) >droYak2.chr3R 20205038 92 - 28832112 ----------------------------AAUGGCAUAAAGUCUUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCAUUUUAAUCGUUUUGAUUAAAAGCCACCACGCCUAAUUAGUUGG ----------------------------...(((.....))).((((((((((((((............(....)..((((((((.....))))))))....)))))))))))))).... ( -28.20, z-score = -4.06, R) >droSec1.super_0 19683920 92 - 21120651 ----------------------------AAUGGCAUAAAGUCUUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCAUUUUAAUCGUUUUGAUUAAAAGGCACCACGCCUAAUUAGUUGG ----------------------------...(((.....))).((((((((((((((.......((....))..((.((((((((.....)))))))).)).)))))))))))))).... ( -29.00, z-score = -4.17, R) >droSim1.chr3R 19140149 92 - 27517382 ----------------------------AAUGGCAUAAAGUCUUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCAUUUUAAUCGUUUUGAUUAAAAGGCACCACGCCUAAUUAGUUGG ----------------------------...(((.....))).((((((((((((((.......((....))..((.((((((((.....)))))))).)).)))))))))))))).... ( -29.00, z-score = -4.17, R) >droVir3.scaffold_13047 8949883 92 - 19223366 ----------------------------AAUGGCAUAAAGUCUUUGAUUAGGCGUGGAAAAAUAUUGUCGGAAAGCAUUUUAAUCGUUUUGAUUAAAAGCCACCACGCCUAAUUAGUUGG ----------------------------...(((.....))).((((((((((((((........(((......)))((((((((.....))))))))....)))))))))))))).... ( -27.60, z-score = -3.69, R) >droMoj3.scaffold_6540 2435917 92 + 34148556 ----------------------------AAUGGCAUAAAGUCUUUGAUUAGGCGUGGAAAAAUAUUGUCGGAAAGCAUUUUAAUCGUUUUGAUUAAAAGCCACCACGCCUAAUUAGUUGG ----------------------------...(((.....))).((((((((((((((........(((......)))((((((((.....))))))))....)))))))))))))).... ( -27.60, z-score = -3.69, R) >droGri2.scaffold_15074 7059633 92 + 7742996 ----------------------------AAUGGUAUUAAGUCUUUGAUUAGGCGUCGAAAAAUAUUGUCGGAAAGCAUUUUAAUCGUUUUGAUUAAAAGCCACCACGCCUAAUUAGUUGG ----------------------------...............(((((((((((((((......)))..((...((.((((((((.....))))))))))..)))))))))))))).... ( -22.10, z-score = -2.18, R) >droWil1.scaffold_181130 9373792 120 - 16660200 UAUAAAACUACAACAACAACCACAUCGAAAAAACCGAAACUCUUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCACUUUAAUCGUUUUGAUUAAAAGCCAGCAAGCCUAAUUAGUUGG .............((((.......(((.......))).......(((((((((.(((.........((.(....).))(((((((.....)))))))..)))....))))))))))))). ( -24.70, z-score = -2.63, R) >consensus ____________________________AAUGGCAUAAAGUCUUUGAUUAGGCGUGGAAAAAUAUUGUCCGAAAGCAUUUUAAUCGUUUUGAUUAAAAGCCACCACGCCUAAUUAGUUGG ...............................(((.....))).((((((((((((((........(((......)))((((((((.....))))))))....)))))))))))))).... (-24.79 = -25.14 + 0.35)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:38:14 2011