| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,300,388 – 19,300,484 |
| Length | 96 |
| Max. P | 0.850194 |

| Location | 19,300,388 – 19,300,484 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 76.94 |
| Shannon entropy | 0.45375 |
| G+C content | 0.45363 |
| Mean single sequence MFE | -26.91 |
| Consensus MFE | -12.32 |
| Energy contribution | -12.68 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.850194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
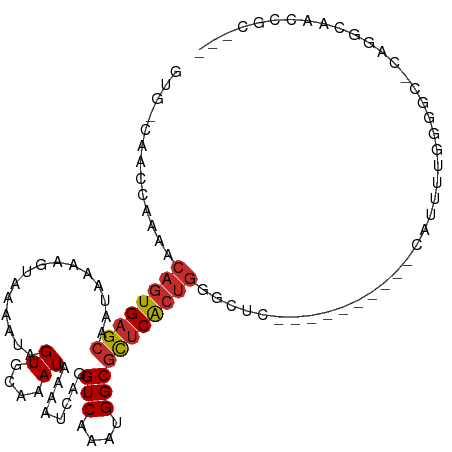
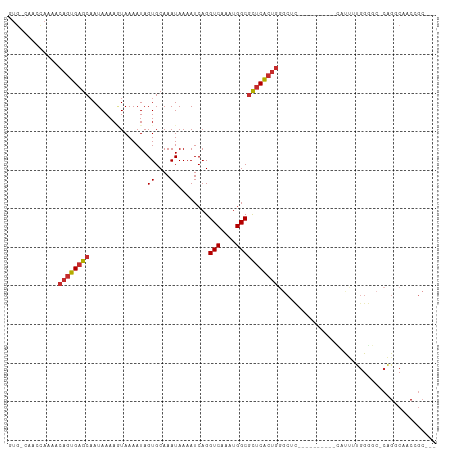
>dm3.chr3R 19300388 96 - 27905053 GUG-CAACUAUAACAGUGAGCAAUAAAAGUAAAAUAGUGCAAAUAAAAUCAGGUCAAAUGGCGCUCACUGGGCUC----------CAUUUUGGGGC-CAGGCAACCGC--- .((-(........((((((((.......(((......)))............(((....)))))))))))(((((----------(.....)))))-)..))).....--- ( -32.40, z-score = -3.66, R) >droSim1.chr3R 19120920 96 - 27517382 GUG-CAACCAUAACAGUGAGCAAUAAAAGUAAAAUAGUGCAAAUAAAAUCAGGUCAAAUGGCGCUCACUGGGCUC----------CAUUUCGGGGC-CAGGCAACCGC--- .((-(........((((((((.......(((......)))............(((....)))))))))))(((((----------(.....)))))-)..))).....--- ( -32.10, z-score = -3.63, R) >droSec1.super_0 19663453 96 - 21120651 GUG-CAACCAUAACAGUGAGCUAUAAAAGUAAAAUAGUGCAAAUAAAAUCAGGUCAAAUGGCGCUCACUGGGCUC----------CAUUUCGGGGC-CAGGCAACCGC--- .((-(........((((((((.......(((......)))............(((....)))))))))))(((((----------(.....)))))-)..))).....--- ( -31.60, z-score = -3.26, R) >droYak2.chr3R 20184135 96 - 28832112 GUG-CAAGCAAAACAGUGAGCAAUAAAAAUAAAAUAGUGCAAAUAAAAUCAGGUCAAAUGGCGCUCACUGCUCUC----------CAUUUUGGGGC-CAGUCAGCCGC--- (.(-(..((....((((((((...............((....))........(((....)))))))))))..(((----------(.....)))).-..))..)))..--- ( -20.70, z-score = -0.13, R) >droEre2.scaffold_4820 1721097 96 + 10470090 GUG-CAAACAAAACAGUGCGCAAUAAAAGUAAAAUAGUGCAAAUAAAAUCAGGUCAAAUGGCGCUCACUGGGCUC----------CAUUUUGGGGC-CAGGCAGCCGC--- .((-(........(((((.((.......(((......)))............(((....))))).)))))(((((----------(.....)))))-)..))).....--- ( -27.90, z-score = -1.53, R) >droAna3.scaffold_13340 3445046 107 + 23697760 GUGUCCUGGAAAACAGUGAGCAAUAAAAGUAAAAUAGUGCAAAUAAAAUCAGGUCAAAUGGCGCUCACUGGGCUCUAAAAAGCAUUAUUUUGGGUC-CAGGCAGCCGC--- .((.((((((...((((((((.......(((......)))............(((....))))))))))).(((......)))...........))-)))))).....--- ( -31.10, z-score = -2.63, R) >dp4.chr2 2785888 91 - 30794189 -----------AACAGUGAGCAAUAAAACUAAAAUAGUGCAAAUAAAAUCAGGUCAAAUGGCGCUCACUGGGCAU---------UGGCCACGGACCACGGACCACGGCCCC -----------..((((((((......((((...))))..............(((....)))))))))))(((..---------(((.(.((.....))).)))..))).. ( -26.90, z-score = -1.99, R) >droPer1.super_7 2879139 91 + 4445127 -----------AACAGUGAGCAAUAAAACUAAAAUAGUGCAAAUAAAAUCAGGUCAAAUGGCGCUCACUGGGCAU---------UGGCCACGGACCACGGACCACGGCCCC -----------..((((((((......((((...))))..............(((....)))))))))))(((..---------(((.(.((.....))).)))..))).. ( -26.90, z-score = -1.99, R) >droWil1.scaffold_181130 9347107 86 - 16660200 --GUGCUUUCGAACAGUGAGCAAUAAAAGUAAAAUAGUGCAAAUAAAAUCAGGUCAAAUGGCAUUCG-----------------AAAUAAUGGUAAGGAGGCAGC------ --.(((((((..((.....(((.((.........)).)))............(((....))).....-----------------........))..)))))))..------ ( -12.60, z-score = -0.28, R) >consensus GUG_CAACCAAAACAGUGAGCAAUAAAAGUAAAAUAGUGCAAAUAAAAUCAGGUCAAAUGGCGCUCACUGGGCUC__________CAUUUUGGGGC_CAGGCAACCGC___ .............((((((((...............((....))........(((....)))))))))))......................................... (-12.32 = -12.68 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:38:08 2011