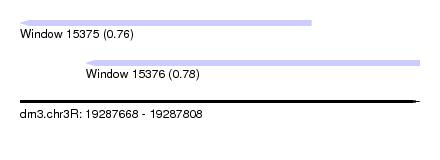
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,287,668 – 19,287,808 |
| Length | 140 |
| Max. P | 0.775148 |

| Location | 19,287,668 – 19,287,770 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.01 |
| Shannon entropy | 0.41708 |
| G+C content | 0.54508 |
| Mean single sequence MFE | -27.93 |
| Consensus MFE | -17.09 |
| Energy contribution | -16.98 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.760197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19287668 102 - 27905053 CUCAUUAAAGUUUUAAACACGUCGCUCACGUAGUGACGCGUCUGUCAGACGGCUGCCAUGUAGUCGAGUAGGCACCCU-CUGCCCGCUCCCACACCCAAUGGC--------- ........(((........(((((((.....)))))))((((.....)))))))(((((...((.(((..((((....-.))))..)))....))...)))))--------- ( -29.70, z-score = -1.41, R) >droSim1.chr3R 19107619 111 - 27517382 CUCAUUAAAGUUUUAAACACGUCGCUCACGUAGUGACGCGUCUGUCAGACGGCUGCCAUGUAGUCGAGUAGGCACCCU-CUGCCCGCUCCCACACCCAAUGGCCAUGUGGUU .........((((((.((.(((((((.....))))))).)).))..))))((((((...))))))(((..((((....-.))))..)))(((((((....))...))))).. ( -33.70, z-score = -1.34, R) >droSec1.super_0 19650766 111 - 21120651 CUCAUUAAAGUUUUAAACACGUCGCUCACGUAGUGACGCGUCUGUCAGACGGCUGCCAUGUAGUCGAGUAGGCACCCU-CUGCCCGCUCCCACACCCAAUGGCUAUGUGGUU ........(((........(((((((.....)))))))((((.....)))))))((((((((((((((..((((....-.))))..)))...........))))))))))). ( -35.80, z-score = -1.99, R) >droYak2.chr3R 20171097 111 - 28832112 CUCAUUAAAGUUUUAAACACGUCGCUCACGUAGUGACGCGUCUGUCAGACGGCUGCCAUGUAGUCGAGUAGGCACCCU-CUGCCCGCUCCCACACCCAAUGGAUCUGUAGUU ................((.(((((((.....))))))).))(((.((((.((((((...))))))(((..((((....-.))))..)))(((.......))).))))))).. ( -32.10, z-score = -1.51, R) >droAna3.scaffold_13340 3431538 100 + 23697760 CUCAUUAAAGUUUUAAACACGUCGCUCACGUAGUGACGCGUCUGUCAGACGGCUGCCAUGUAGCC----CGGCACCCU-CUUCCCGCUCCCACACCCAACAAUUC------- .........(((........((((((.....))))))(((((.....)))((((((...))))))----..)).....-..................))).....------- ( -20.20, z-score = -0.83, R) >droWil1.scaffold_181130 9332591 101 - 16660200 CUCAUUAAACUUUUAAACACGUCGUUCACGUAUUGACGCGUCUGUCAGACGGCUGCCAUGUAGUC----AGGCACCCUGCCCCCUUCGUCCCCUCUAAACCGCCU------- ................((.(((((.........))))).))......(((((((((...))))))----.(((.....)))......)))...............------- ( -18.40, z-score = -0.40, R) >droVir3.scaffold_13047 8920084 85 - 19223366 CUCAUUAAACUUUUAAACACGUCGCUCACGUAGUGACGCGUCUGUCAGACGGCUGCCAUGUAGCC----A-ACG--CUGGCUGCCGCAUCCC-------------------- ....................((((((.....))))))(((.(.(((((..((((((...))))))----.-...--))))).).))).....-------------------- ( -26.90, z-score = -2.05, R) >droMoj3.scaffold_6540 2402770 85 + 34148556 CUCAUUAAACUUUUAAACACGUCGCUCACGUAGUGACGCGUCUGUCAGAUGGCUGCCAUGUAGCC----A-GCA--CUGGUCGCCGCAUCCC-------------------- ....................((((((.....))))))(((.(..((((.(((((((...))))))----)-...--))))..).))).....-------------------- ( -25.80, z-score = -1.86, R) >droGri2.scaffold_15074 7030295 86 + 7742996 CUCAUUAAACUUUUAAACACGUCGCUCACGUAGUGACGCGUCUGUCAGACGGCUGCCAUGUAGCC----AGGCA--AAGGCAGCCGCAUCCC-------------------- .........((..((.((.(((((((.....))))))).)).))..)).((((((((.(((....----..)))--..))))))))......-------------------- ( -28.80, z-score = -2.60, R) >consensus CUCAUUAAAGUUUUAAACACGUCGCUCACGUAGUGACGCGUCUGUCAGACGGCUGCCAUGUAGUC____AGGCACCCU_CUGCCCGCUCCCACACCCAAUGGC_________ ................((.(((((((.....))))))).)).((((....((((((...)))))).....))))...................................... (-17.09 = -16.98 + -0.11)
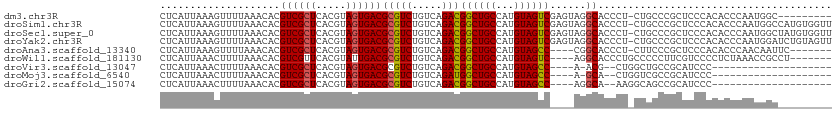

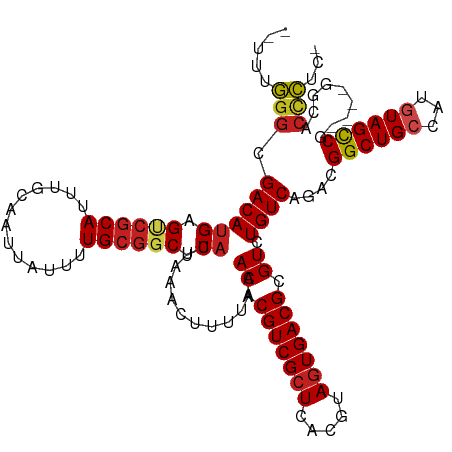
| Location | 19,287,691 – 19,287,808 |
|---|---|
| Length | 117 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.48 |
| Shannon entropy | 0.19851 |
| G+C content | 0.50179 |
| Mean single sequence MFE | -38.43 |
| Consensus MFE | -30.26 |
| Energy contribution | -30.19 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.775148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19287691 117 - 27905053 --UUUGGGCGACAUGAGUCGCAUUUACAAUUAUUUUGCGGCUCAUUAAAGUUUUAAACACGUCGCUCACGUAGUGACGCGUCUGUCAGACGGCUGCCAUGUAGUCGAGUAGGCACCCUC- --...(((.(((((((((((((.............))))))))))..............(((((((.....))))))).)))((((...(((((((...)))))))....)))))))..- ( -39.52, z-score = -2.56, R) >droSim1.chr3R 19107651 117 - 27517382 --UUUGGGCGACAUGAGUCGCAUUUGCAAUUAUUUUGCGGCUCAUUAAAGUUUUAAACACGUCGCUCACGUAGUGACGCGUCUGUCAGACGGCUGCCAUGUAGUCGAGUAGGCACCCUC- --...(((.(((((((((((((.............))))))))))..............(((((((.....))))))).)))((((...(((((((...)))))))....)))))))..- ( -39.52, z-score = -2.11, R) >droSec1.super_0 19650798 117 - 21120651 --UUUGGGCGACAUGAGUCGCAUUUGCAAUUAUUUUGAGGCUCAUUAAAGUUUUAAACACGUCGCUCACGUAGUGACGCGUCUGUCAGACGGCUGCCAUGUAGUCGAGUAGGCACCCUC- --...(((((((....)))))....((......(((((((((......)))))))))...((((((.....))))))))(((((.(...(((((((...))))))).))))))..))..- ( -34.80, z-score = -0.91, R) >droYak2.chr3R 20171129 117 - 28832112 --UUUGGGCGACAUGAGUCGCAUUUGCAAUUAUUUUGCGGCUCAUUAAAGUUUUAAACACGUCGCUCACGUAGUGACGCGUCUGUCAGACGGCUGCCAUGUAGUCGAGUAGGCACCCUC- --...(((.(((((((((((((.............))))))))))..............(((((((.....))))))).)))((((...(((((((...)))))))....)))))))..- ( -39.52, z-score = -2.11, R) >droEre2.scaffold_4820 1708335 116 + 10470090 --UUUGGGCGACAUGAGUAGCAUUUGCAAUUAUUUUGCGGCUCAUUAAAGUUUUAAACACGUCGCUCACGUAGUGACGCGUCUGUCAGACGGCUGCCAUGUAGUCGAGU-GGCACCCUC- --...(((.(((((((((.(((.............))).))))))..............(((((((.....))))))).)))(((((..(((((((...)))))))..)-)))))))..- ( -37.92, z-score = -1.65, R) >droAna3.scaffold_13340 3431563 113 + 23697760 --UUUGGGCGACACGAGUCGCAUUUGCAAUUAUUUUGCGGCUCAUUAAAGUUUUAAACACGUCGCUCACGUAGUGACGCGUCUGUCAGACGGCUGCCAUGUAGCCC----GGCACCCUC- --..((((((((..((((((((.............))))))))......((.....))..))))))))....(((.((.(((.....)))((((((...)))))))----).)))....- ( -39.02, z-score = -2.17, R) >droPer1.super_7 2864649 110 + 4445127 -----GGGCGACAUGAGGCGCAUUUGCAAUUAUUUUGCGGCUCAUUAAACUUUUAAACACGUCGCUCACGUAGUGACGCGUCUGUCAGACGGCUGGCAUGUAGCCG----AGCACCCCA- -----(((.((((((((.((((.............)))).))))............((.(((((((.....))))))).)).))))...((((((.....))))))----.....))).- ( -38.42, z-score = -1.80, R) >dp4.chr2 2771422 110 - 30794189 -----GGGCGACAUGAGGCGCAUUUGCAAUUAUUUUGCGGCUCAUUAAACUUUUAAACACGUCGCUCACGUAGUGACGCGUCUGUCAGACGGCUGGCAUGUAGCCG----GGCACCCCA- -----(((.((((((((.((((.............)))).))))............((.(((((((.....))))))).)).))))....(.(((((.....))))----).)..))).- ( -40.12, z-score = -2.05, R) >droWil1.scaffold_181130 9332616 116 - 16660200 UUUUUGGGCGACAUGAGUCGCAUUUGCAAUUAUUUUGCGGCUCAUUAAACUUUUAAACACGUCGUUCACGUAUUGACGCGUCUGUCAGACGGCUGCCAUGUAGUCA----GGCACCCUGC ....((((((((.(((((((((.............)))))))))((((....))))....)))))))).((.((((((....))))))))((.((((.((....))----)))).))... ( -36.62, z-score = -2.13, R) >droVir3.scaffold_13047 8920095 112 - 19223366 --UUUUGGCGACAUGCGCCGCAUUUGCAAUUAUUUUGCGGCUCAUUAAACUUUUAAACACGUCGCUCACGUAGUGACGCGUCUGUCAGACGGCUGCCAUGUAGCCA-----ACGCUGGC- --..(..((((((.((((((((.............))))))..................(((((((.....))))))).)).))))....((((((...)))))).-----..))..).- ( -38.72, z-score = -1.85, R) >droMoj3.scaffold_6540 2402781 112 + 34148556 --UUUUGGCGACAUGCGCCGCAUUUGCAAUUAUUUUGCGGCUCAUUAAACUUUUAAACACGUCGCUCACGUAGUGACGCGUCUGUCAGAUGGCUGCCAUGUAGCCA-----GCACUGGU- --.((((((((((((.((((((.............)))))).)))..............(((((((.....))))))).))).))))))(((((((...)))))))-----........- ( -38.52, z-score = -1.90, R) >droGri2.scaffold_15074 7030306 113 + 7742996 --UUUUGGCGACAUGCGCCGCAUUUGCAAUUAUUUUGCGGCUCAUUAAACUUUUAAACACGUCGCUCACGUAGUGACGCGUCUGUCAGACGGCUGCCAUGUAGCCA----GGCAAAGGC- --(((((.(((((.((((((((.............))))))..................(((((((.....))))))).)).))))....((((((...)))))).----).)))))..- ( -38.42, z-score = -1.64, R) >consensus __UUUGGGCGACAUGAGUCGCAUUUGCAAUUAUUUUGCGGCUCAUUAAACUUUUAAACACGUCGCUCACGUAGUGACGCGUCUGUCAGACGGCUGCCAUGUAGCCG____GGCACCCUC_ .....(((.((((((.((((((.............)))))).))............((.(((((((.....))))))).)).))))....((((((...)))))).........)))... (-30.26 = -30.19 + -0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:38:06 2011