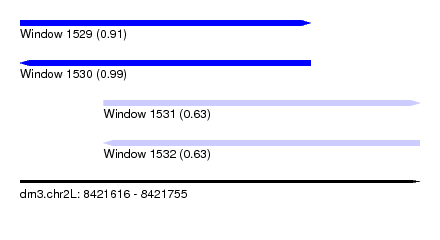
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,421,616 – 8,421,755 |
| Length | 139 |
| Max. P | 0.991814 |

| Location | 8,421,616 – 8,421,717 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.09 |
| Shannon entropy | 0.36276 |
| G+C content | 0.41631 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -11.84 |
| Energy contribution | -11.44 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8421616 101 + 23011544 ---AAAGUACGAGAAUAUCUUGAAGAU----AUGAUAAAAACCGGCUA-GAUAAGCCGGUCGUAUCUGUAUCUG------CUGUCAUGUUGUGAAAAACUUGACUGGAGCACGUC- ---......((((.....)))).((((----((((((...(((((((.-....)))))))..)))).))))))(------((((((.(((......))).))))...))).....- ( -31.30, z-score = -4.01, R) >droSim1.chr2L 8207853 107 + 22036055 ---AUGGUACGAGAAUAUCUUGAAGAU----AUGAUAAAAACCGGCUA-GAUAAGCCGUUCGUAUCUGUAUCUGUAUCUGCUGUCAUGUUGUGAAAAACUUGACUGGAGCACGUC- ---..((((((((.....)))..((((----((((((..(((.((((.-....)))))))..)))).)))))))))))((((((((.(((......))).))))...))))....- ( -29.30, z-score = -2.65, R) >droSec1.super_3 3918050 101 + 7220098 ---AAAGUACGAGAAUAUCUUGAAGAU----AUGAUAAAAACCGGCUA-GAUAAGCCGGUCGUAUCUGUAUCUA------CUGUCAUGUUGUGAAAAACUUGACUGGAGCACGUC- ---..(((.((((.....)))).((((----((((((...(((((((.-....)))))))..)))).)))))))------))((((.(((......))).))))...........- ( -29.60, z-score = -3.55, R) >droYak2.chr2L 11059470 101 + 22324452 ---AAAGUACGAGAAUAUCUUGAAGAU----AUGAUAAAAACCGGCUA-GAUAAGCUGGUCGUAUCUGUAUCUG------CUGUCAUGUUGUGAAAAACUUGACUGGAGCACGUC- ---......((((.....)))).((((----((((((...(((((((.-....)))))))..)))).))))))(------((((((.(((......))).))))...))).....- ( -29.10, z-score = -3.37, R) >droEre2.scaffold_4929 17330321 101 + 26641161 ---AAAGUACGAGAAUAUCUUGAAGAU----AUGAUAAAAACCGGCUA-GAUAAGCCGGUCGUAUCUGUAUCUG------CUGCCAUGUUGUGAAAAACUUGACUGGAGCACGUC- ---......((((.....)))).((((----((((((...(((((((.-....)))))))..)))).))))))(------((.(((.(((((.....))..))))))))).....- ( -30.80, z-score = -3.91, R) >droAna3.scaffold_12916 10078048 89 + 16180835 -AGUUCGAGUAAGUAUAUCUUCAAGAU----AUGAUAAAAACCGGCUAAGAUAAGCCG-----------GUCUG-----------UUGCUCUGGAAAACUUGACUGCAGCACGUCU -..(((((((((.((((((.....)))----)))......(((((((......)))))-----------))...-----------)))))).)))......(((........))). ( -23.50, z-score = -1.82, R) >dp4.chr4_group1 1189903 99 - 5278887 AGUGGAGUACAAGAAUAUCUUGAAGAUGGGAACGAUAAAAACCGGCUA-GAUAAGCCG---------GUAUCUA-----GCUGUCAGGCAGUGAAAA-CUUGACUAGAGCACGUC- (((.((((.((((.....)))).(((((....).......(((((((.-....)))))---------)))))).-----((((.....))))....)-))).)))..........- ( -27.00, z-score = -2.13, R) >droPer1.super_5 1175819 99 + 6813705 AGUGGAGUACAAGAAUAUCUUGAAGAUGGGAACGAUGAAAACCGGCUA-GAUAAGCCG---------GUAUCUA-----GCUGUCAGGCAGUGAAAA-CUUGACUAGAGCACGUC- (((.((((.((((.....)))).(((((....).......(((((((.-....)))))---------)))))).-----((((.....))))....)-))).)))..........- ( -27.00, z-score = -1.92, R) >consensus ___AAAGUACGAGAAUAUCUUGAAGAU____AUGAUAAAAACCGGCUA_GAUAAGCCGGUCGUAUCUGUAUCUG______CUGUCAUGUUGUGAAAAACUUGACUGGAGCACGUC_ .....(((.((((..((((..............)))).....(((((......)))))........................................)))))))........... (-11.84 = -11.44 + -0.40)
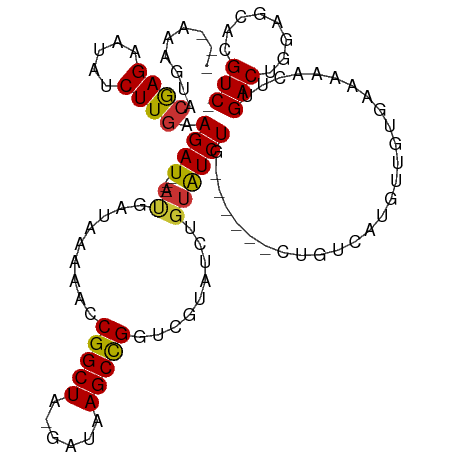
| Location | 8,421,616 – 8,421,717 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.09 |
| Shannon entropy | 0.36276 |
| G+C content | 0.41631 |
| Mean single sequence MFE | -25.24 |
| Consensus MFE | -12.72 |
| Energy contribution | -13.81 |
| Covariance contribution | 1.10 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.991814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8421616 101 - 23011544 -GACGUGCUCCAGUCAAGUUUUUCACAACAUGACAG------CAGAUACAGAUACGACCGGCUUAUC-UAGCCGGUUUUUAUCAU----AUCUUCAAGAUAUUCUCGUACUUU--- -...((((....((((.(((......))).)))).(------.(((((..((((.((((((((....-.))))))))..)))).)----)))).)...........))))...--- ( -28.50, z-score = -4.73, R) >droSim1.chr2L 8207853 107 - 22036055 -GACGUGCUCCAGUCAAGUUUUUCACAACAUGACAGCAGAUACAGAUACAGAUACGAACGGCUUAUC-UAGCCGGUUUUUAUCAU----AUCUUCAAGAUAUUCUCGUACCAU--- -....((((...((((.(((......))).))))))))(.((((((....((((.((((((((....-.)))).)))).))))((----(((.....)))))))).))).)..--- ( -24.50, z-score = -2.89, R) >droSec1.super_3 3918050 101 - 7220098 -GACGUGCUCCAGUCAAGUUUUUCACAACAUGACAG------UAGAUACAGAUACGACCGGCUUAUC-UAGCCGGUUUUUAUCAU----AUCUUCAAGAUAUUCUCGUACUUU--- -...((((....((((.(((......))).)))).(------.(((((..((((.((((((((....-.))))))))..)))).)----)))).)...........))))...--- ( -28.50, z-score = -4.58, R) >droYak2.chr2L 11059470 101 - 22324452 -GACGUGCUCCAGUCAAGUUUUUCACAACAUGACAG------CAGAUACAGAUACGACCAGCUUAUC-UAGCCGGUUUUUAUCAU----AUCUUCAAGAUAUUCUCGUACUUU--- -...((((....((((.(((......))).)))).(------.(((((..((((.((((.(((....-.))).))))..)))).)----)))).)...........))))...--- ( -22.40, z-score = -3.01, R) >droEre2.scaffold_4929 17330321 101 - 26641161 -GACGUGCUCCAGUCAAGUUUUUCACAACAUGGCAG------CAGAUACAGAUACGACCGGCUUAUC-UAGCCGGUUUUUAUCAU----AUCUUCAAGAUAUUCUCGUACUUU--- -...((((....((((.(((......))).)))).(------.(((((..((((.((((((((....-.))))))))..)))).)----)))).)...........))))...--- ( -27.90, z-score = -4.17, R) >droAna3.scaffold_12916 10078048 89 - 16180835 AGACGUGCUGCAGUCAAGUUUUCCAGAGCAA-----------CAGAC-----------CGGCUUAUCUUAGCCGGUUUUUAUCAU----AUCUUGAAGAUAUACUUACUCGAACU- .(((........)))..((((((.(((....-----------.((((-----------(((((......))))))))).......----.))).))))))...............- ( -23.02, z-score = -2.25, R) >dp4.chr4_group1 1189903 99 + 5278887 -GACGUGCUCUAGUCAAG-UUUUCACUGCCUGACAGC-----UAGAUAC---------CGGCUUAUC-UAGCCGGUUUUUAUCGUUCCCAUCUUCAAGAUAUUCUUGUACUCCACU -((((.(((...((((.(-(.......)).)))))))-----((((.((---------(((((....-.))))))).)))).))))........((((.....))))......... ( -23.70, z-score = -2.48, R) >droPer1.super_5 1175819 99 - 6813705 -GACGUGCUCUAGUCAAG-UUUUCACUGCCUGACAGC-----UAGAUAC---------CGGCUUAUC-UAGCCGGUUUUCAUCGUUCCCAUCUUCAAGAUAUUCUUGUACUCCACU -((((.(((...((((.(-(.......)).)))))))-----..((.((---------(((((....-.)))))))..))..))))........((((.....))))......... ( -23.40, z-score = -2.34, R) >consensus _GACGUGCUCCAGUCAAGUUUUUCACAACAUGACAG______CAGAUACAGAUACGACCGGCUUAUC_UAGCCGGUUUUUAUCAU____AUCUUCAAGAUAUUCUCGUACUUU___ ....((((....((((.(((......))).))))........................(((((......)))))................................))))...... (-12.72 = -13.81 + 1.10)

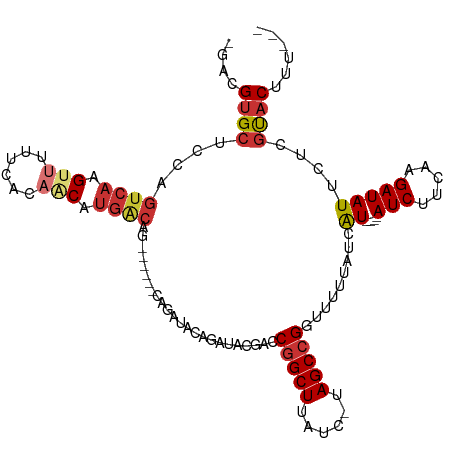
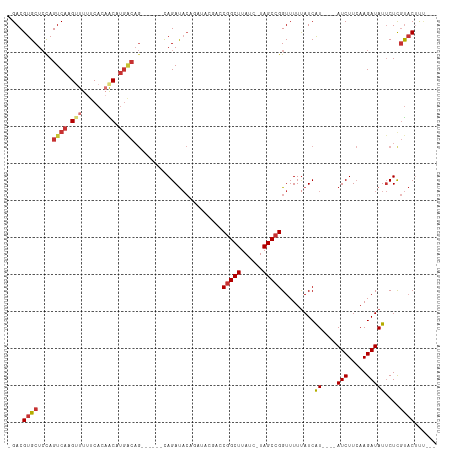
| Location | 8,421,645 – 8,421,755 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.00 |
| Shannon entropy | 0.29672 |
| G+C content | 0.43059 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -23.04 |
| Energy contribution | -23.28 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.23 |
| Mean z-score | -0.84 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.629519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8421645 110 + 23011544 AAAAACCGGCUA-GAUAAGCCGGUCGUAUCUGUAUCUG------CUGUCAUGUUGUGAAAAACUUGACUGGAGCACGUCUCCAC--AAUGUGUACACUCU-GCAAGUUCUUAUUUUGUGG ....(((((((.-....)))))))......(((((..(------(.((((.(((......))).))))(((((.....))))).--...)))))))..(.-.((((.......))))..) ( -28.00, z-score = -1.24, R) >droSim1.chr2L 8207882 116 + 22036055 AAAAACCGGCUA-GAUAAGCCGUUCGUAUCUGUAUCUGUAUCUGCUGUCAUGUUGUGAAAAACUUGACUGGAGCACGUCUCCAC--AAUGUGUACACUCU-GCAAGUUCUUAUUUCGUGG ......(((((.-....)))))..........................((((..((((..((((((...(((((((((......--.)))))....))))-.)))))).))))..)))). ( -23.20, z-score = 0.21, R) >droSec1.super_3 3918079 110 + 7220098 AAAAACCGGCUA-GAUAAGCCGGUCGUAUCUGUAUCUA------CUGUCAUGUUGUGAAAAACUUGACUGGAGCACGUCUCCAC--AAUGUGUACACUCU-GCAAGUUCUUAUUUCGUGG ....(((((((.-....)))))))..............------....((((..((((..((((((...(((((((((......--.)))))....))))-.)))))).))))..)))). ( -27.70, z-score = -1.38, R) >droYak2.chr2L 11059499 110 + 22324452 AAAAACCGGCUA-GAUAAGCUGGUCGUAUCUGUAUCUG------CUGUCAUGUUGUGAAAAACUUGACUGGAGCACGUCUCCAC--AAUGUGUACACUCU-GCAAGUUCUUAUUUCGUGG ....(((((((.-....)))))))..............------....((((..((((..((((((...(((((((((......--.)))))....))))-.)))))).))))..)))). ( -25.50, z-score = -0.49, R) >droEre2.scaffold_4929 17330350 110 + 26641161 AAAAACCGGCUA-GAUAAGCCGGUCGUAUCUGUAUCUG------CUGCCAUGUUGUGAAAAACUUGACUGGAGCACGUCUCCAC--AAUGUGUACACUCU-GCAAGUUCUUAUUUCGUGG ....(((((((.-....)))))))..............------...(((((..((((..((((((...(((((((((......--.)))))....))))-.)))))).))))..))))) ( -29.80, z-score = -1.67, R) >droAna3.scaffold_12916 10078079 90 + 16180835 AAAAACCGGCUAAGAUAAGCCGGUC-----UGU-----------------UGCUCUGGAAAACUUGACUGCAGCACGUCUUUUCUUAAUGUUUAAAUUUCAGUCAGUUACUA-------- ....(((((((......))))))).-----(((-----------------(((..(.((....)).)..)))))).....................................-------- ( -18.20, z-score = -0.46, R) >consensus AAAAACCGGCUA_GAUAAGCCGGUCGUAUCUGUAUCUG______CUGUCAUGUUGUGAAAAACUUGACUGGAGCACGUCUCCAC__AAUGUGUACACUCU_GCAAGUUCUUAUUUCGUGG ....(((((((......))))))).......................(((((..((((..((((((...(((((((((.........)))))....))))..)))))).))))..))))) (-23.04 = -23.28 + 0.24)

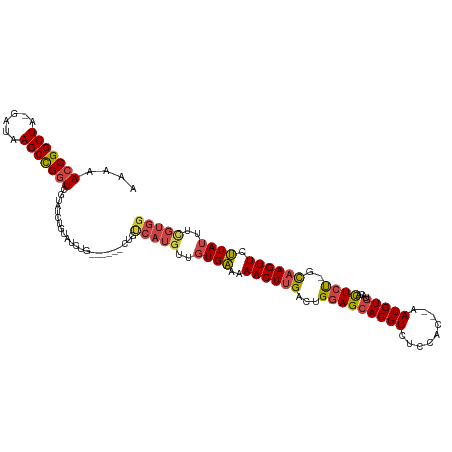
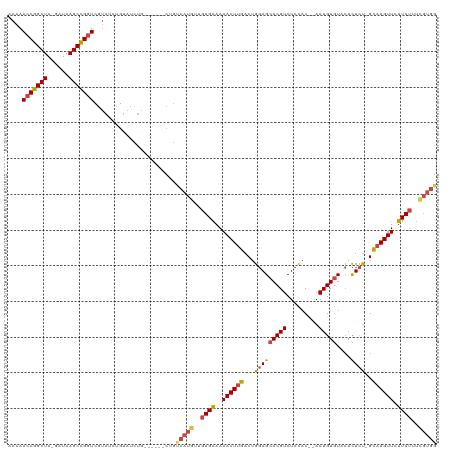
| Location | 8,421,645 – 8,421,755 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.00 |
| Shannon entropy | 0.29672 |
| G+C content | 0.43059 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -19.88 |
| Energy contribution | -21.27 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.629744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8421645 110 - 23011544 CCACAAAAUAAGAACUUGC-AGAGUGUACACAUU--GUGGAGACGUGCUCCAGUCAAGUUUUUCACAACAUGACAG------CAGAUACAGAUACGACCGGCUUAUC-UAGCCGGUUUUU ...............((((-..((((....))))--.(((((.....)))))((((.(((......))).)))).)------)))..........((((((((....-.))))))))... ( -28.90, z-score = -2.19, R) >droSim1.chr2L 8207882 116 - 22036055 CCACGAAAUAAGAACUUGC-AGAGUGUACACAUU--GUGGAGACGUGCUCCAGUCAAGUUUUUCACAACAUGACAGCAGAUACAGAUACAGAUACGAACGGCUUAUC-UAGCCGGUUUUU .........((((((((((-..((((....))))--.(((((.....)))))((((.(((......))).)))).))))...................(((((....-.))))))))))) ( -26.20, z-score = -0.99, R) >droSec1.super_3 3918079 110 - 7220098 CCACGAAAUAAGAACUUGC-AGAGUGUACACAUU--GUGGAGACGUGCUCCAGUCAAGUUUUUCACAACAUGACAG------UAGAUACAGAUACGACCGGCUUAUC-UAGCCGGUUUUU ...................-...((((..((...--.(((((.....)))))((((.(((......))).)))).)------)..))))......((((((((....-.))))))))... ( -28.80, z-score = -1.95, R) >droYak2.chr2L 11059499 110 - 22324452 CCACGAAAUAAGAACUUGC-AGAGUGUACACAUU--GUGGAGACGUGCUCCAGUCAAGUUUUUCACAACAUGACAG------CAGAUACAGAUACGACCAGCUUAUC-UAGCCGGUUUUU .........(((((((.((-...((((.......--.(((((.....)))))((((.(((......))).))))..------...))))(((((.(.....).))))-).)).))))))) ( -23.20, z-score = -0.50, R) >droEre2.scaffold_4929 17330350 110 - 26641161 CCACGAAAUAAGAACUUGC-AGAGUGUACACAUU--GUGGAGACGUGCUCCAGUCAAGUUUUUCACAACAUGGCAG------CAGAUACAGAUACGACCGGCUUAUC-UAGCCGGUUUUU (((......((((((((((-..((((....))))--.(((((.....)))))).))))))))).......)))...------.............((((((((....-.))))))))... ( -29.22, z-score = -1.67, R) >droAna3.scaffold_12916 10078079 90 - 16180835 --------UAGUAACUGACUGAAAUUUAAACAUUAAGAAAAGACGUGCUGCAGUCAAGUUUUCCAGAGCA-----------------ACA-----GACCGGCUUAUCUUAGCCGGUUUUU --------.......((((((........((.((.......)).))....)))))).((((....)))).-----------------..(-----((((((((......))))))))).. ( -20.30, z-score = -1.30, R) >consensus CCACGAAAUAAGAACUUGC_AGAGUGUACACAUU__GUGGAGACGUGCUCCAGUCAAGUUUUUCACAACAUGACAG______CAGAUACAGAUACGACCGGCUUAUC_UAGCCGGUUUUU .........(((((((((....((((....))))...(((((.....)))))..)))))))))................................((((((((......))))))))... (-19.88 = -21.27 + 1.39)

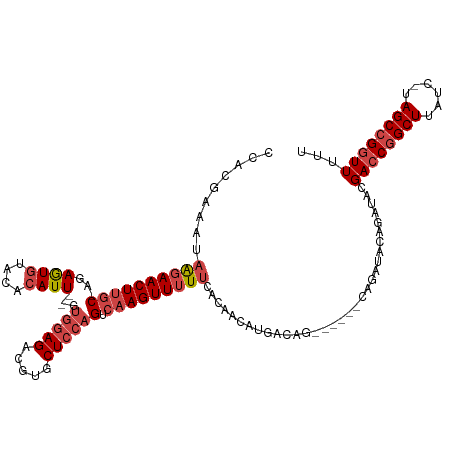
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:25:59 2011