
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,284,755 – 19,284,862 |
| Length | 107 |
| Max. P | 0.775121 |

| Location | 19,284,755 – 19,284,862 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 130 |
| Reading direction | forward |
| Mean pairwise identity | 60.59 |
| Shannon entropy | 0.70093 |
| G+C content | 0.39540 |
| Mean single sequence MFE | -22.05 |
| Consensus MFE | -9.40 |
| Energy contribution | -9.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.775121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
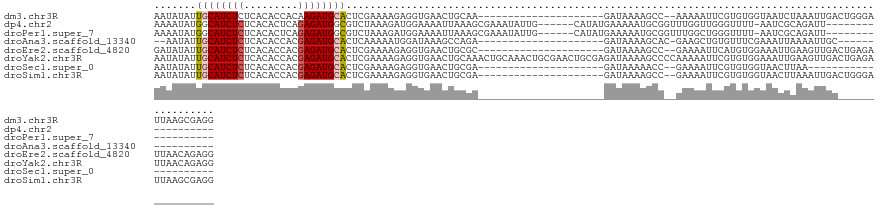
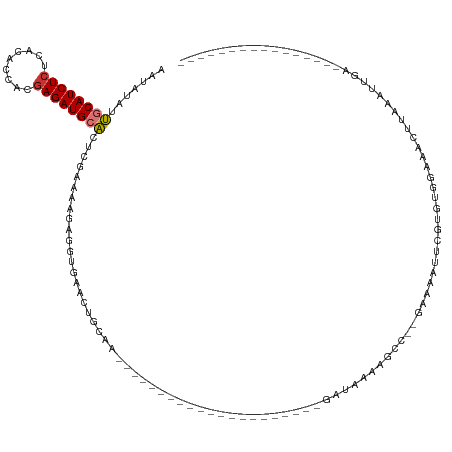
>dm3.chr3R 19284755 107 + 27905053 AAUAUAUUGCAUCUCUCACACCACAAGAUGCACUCGAAAAGAGGUGAACUGCAA---------------------GAUAAAAGCC--AAAAAUUCGUGUGGUAAUCUAAAUUGACUGGGAUUAAGCGAGG ......((((...(((((.((((((.(((((((((.....)))(....))))).---------------------(........)--......)).))))))..((......)).)))))....)))).. ( -19.00, z-score = 0.15, R) >dp4.chr2 2768224 105 + 30794189 AAAAUAUGGCAUCUCUCACACUCAGAGAUGGCGUCUAAAGAUGGAAAAUUAAAGCGAAAUAUUG------CAUAUGAAAAAUGCGGUUUGGUUGGGUUUU-AAUCGCAGAUU------------------ .......(.(((((((.......))))))).)((((...(((..(((((..((.(((...((((------(((.......)))))))))).))..)))))-.)))..)))).------------------ ( -24.00, z-score = -1.52, R) >droPer1.super_7 2861441 105 - 4445127 AAAAUAUGGCAUCUCUCACACUCAGAGAUGGCGUCUAAAGAUGGAAAAUUAAAGCGAAAUAUUG------CAUAUGAAAAAUGCGGUUUGGCUGGGUUUU-AAUCGCAGAUU------------------ .......(.(((((((.......))))))).)((((...(((..(((((...(((.((((...(------(((.......)))).)))).)))..)))))-.)))..)))).------------------ ( -25.80, z-score = -1.92, R) >droAna3.scaffold_13340 3428462 90 - 23697760 --AAUAUUGCAUCUCUCACACCACGAGAUGCACUCAAAAAUGGAUAAAGCCAGA---------------------GAUAAAAGCAC-GAAGCUGUGUUUCGAAAUUAAAAUUGC---------------- --.....((((((((.........)))))))).((.....(((......)))((---------------------((((..(((..-...))).))))))))............---------------- ( -19.70, z-score = -2.07, R) >droEre2.scaffold_4820 1705384 107 - 10470090 GAUAUAUUGCAUCUCUCACACCACGAGAUGCACUCGAAAAGAGGUGAACUGCGC---------------------GAUAAAAGCC--GAAAAUUCAUGUGGAAAUUGAAGUUGACUGAGAUUAACAGAGG .......((((((((.........))))))))(((.......(((.((((...(---------------------(((.....((--(..........)))..)))).)))).)))..........))). ( -21.73, z-score = -0.51, R) >droYak2.chr3R 20168000 130 + 28832112 AAUAUAUUGCAUCUCUCACACCACGAGAUGCACUCGAAAAGAGGUGAACUGCAAACUGCAAACUGCGAACUGCGAGAUAAAAGCCCCAAAAAUUCGUGUGGAAAUUGAAGUUGACUGAGAUUAACAGAGG .......((((((((.........))))))))(((.......(((.((((.(((..(((.....)))..(..((.((................)).))..)...))).)))).)))..........))). ( -26.02, z-score = -0.41, R) >droSec1.super_0 19647894 86 + 21120651 AAUAUAUUGCAUCUCUCACACCACGAGAUGCACUCGAAAAGAGGUGAACUGCGA---------------------GAUAAAAACC--GAAAAUUCGUGUGGUAACUUAA--------------------- .......((((((((.........))))))))(((.....)))((..((..((.---------------------((........--......)).))..)).))....--------------------- ( -18.14, z-score = -1.47, R) >droSim1.chr3R 19104714 107 + 27517382 AAUAUAUUGCAUCUCUCACACCACGAGAUGCACUCGAAAAGAGGUGAACUGCGA---------------------GAUAAAAGCC--GAAAAUUCGUGUGGUAACUUAAAUUGACUGGGAUUAAGCGAGG .......((((((((.........))))))))((((......((....)).)))---------------------).........--.....(((((.((((..(...........)..)))).))))). ( -22.00, z-score = -0.34, R) >consensus AAUAUAUUGCAUCUCUCACACCACGAGAUGCACUCGAAAAGAGGUGAACUGCAA_____________________GAUAAAAGCC__GAAAAUUCGUGUGGAAACUUAAAUUGA________________ .......((((((((.........)))))))).................................................................................................. ( -9.40 = -9.40 + 0.00)

| Location | 19,284,755 – 19,284,862 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 130 |
| Reading direction | reverse |
| Mean pairwise identity | 60.59 |
| Shannon entropy | 0.70093 |
| G+C content | 0.39540 |
| Mean single sequence MFE | -19.35 |
| Consensus MFE | -10.70 |
| Energy contribution | -10.46 |
| Covariance contribution | -0.23 |
| Combinations/Pair | 1.25 |
| Mean z-score | -0.39 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.734371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
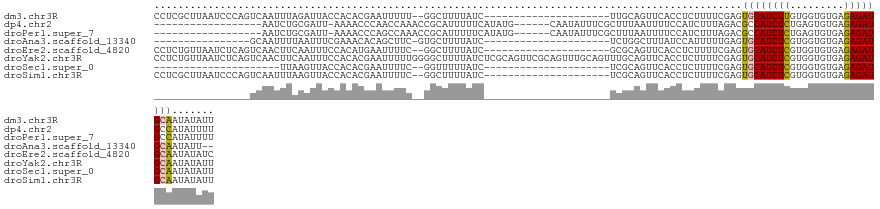
>dm3.chr3R 19284755 107 - 27905053 CCUCGCUUAAUCCCAGUCAAUUUAGAUUACCACACGAAUUUUU--GGCUUUUAUC---------------------UUGCAGUUCACCUCUUUUCGAGUGCAUCUUGUGGUGUGAGAGAUGCAAUAUAUU .(((((........((((......))))((((((.((......--.((.......---------------------..)).((....(((.....))).)).)).))))))))))).............. ( -19.10, z-score = 0.12, R) >dp4.chr2 2768224 105 - 30794189 ------------------AAUCUGCGAUU-AAAACCCAACCAAACCGCAUUUUUCAUAUG------CAAUAUUUCGCUUUAAUUUUCCAUCUUUAGACGCCAUCUCUGAGUGUGAGAGAUGCCAUAUUUU ------------------...........-................((((((((((((((------(........))...............(((((.......))))))))))))))))))........ ( -17.80, z-score = -1.76, R) >droPer1.super_7 2861441 105 + 4445127 ------------------AAUCUGCGAUU-AAAACCCAGCCAAACCGCAUUUUUCAUAUG------CAAUAUUUCGCUUUAAUUUUCCAUCUUUAGACGCCAUCUCUGAGUGUGAGAGAUGCCAUAUUUU ------------------...(((.(...-.....)))).......((((((((((((((------(........))...............(((((.......))))))))))))))))))........ ( -17.90, z-score = -1.31, R) >droAna3.scaffold_13340 3428462 90 + 23697760 ----------------GCAAUUUUAAUUUCGAAACACAGCUUC-GUGCUUUUAUC---------------------UCUGGCUUUAUCCAUUUUUGAGUGCAUCUCGUGGUGUGAGAGAUGCAAUAUU-- ----------------((............(((((((......-))).))))(((---------------------(((.((.....((((....(((.....))))))).)).))))))))......-- ( -18.50, z-score = -0.28, R) >droEre2.scaffold_4820 1705384 107 + 10470090 CCUCUGUUAAUCUCAGUCAACUUCAAUUUCCACAUGAAUUUUC--GGCUUUUAUC---------------------GCGCAGUUCACCUCUUUUCGAGUGCAUCUCGUGGUGUGAGAGAUGCAAUAUAUC ....(((...((((((((...((((.........)))).....--)))...((((---------------------(((.((..(((.((.....)))))...))))))))))))))...)))....... ( -19.40, z-score = 0.10, R) >droYak2.chr3R 20168000 130 - 28832112 CCUCUGUUAAUCUCAGUCAACUUCAAUUUCCACACGAAUUUUUGGGGCUUUUAUCUCGCAGUUCGCAGUUUGCAGUUUGCAGUUCACCUCUUUUCGAGUGCAUCUCGUGGUGUGAGAGAUGCAAUAUAUU ....(((...(((((((.((((.(((........((((((..(((((......))))).))))))....))).)))).))....((((......((((.....)))).)))))))))...)))....... ( -26.40, z-score = 0.41, R) >droSec1.super_0 19647894 86 - 21120651 ---------------------UUAAGUUACCACACGAAUUUUC--GGUUUUUAUC---------------------UCGCAGUUCACCUCUUUUCGAGUGCAUCUCGUGGUGUGAGAGAUGCAAUAUAUU ---------------------.............(((....))--).....((((---------------------((.((...((((......((((.....)))).)))))).))))))......... ( -16.10, z-score = -0.13, R) >droSim1.chr3R 19104714 107 - 27517382 CCUCGCUUAAUCCCAGUCAAUUUAAGUUACCACACGAAUUUUC--GGCUUUUAUC---------------------UCGCAGUUCACCUCUUUUCGAGUGCAUCUCGUGGUGUGAGAGAUGCAAUAUAUU .(((((((((...........))))))........(((((..(--((........---------------------))).)))))..........)))((((((((.........))))))))....... ( -19.60, z-score = -0.29, R) >consensus ________________UCAAUUUAAAUUUCCACACGAAUUUUC__GGCUUUUAUC_____________________UCGCAGUUCACCUCUUUUCGAGUGCAUCUCGUGGUGUGAGAGAUGCAAUAUAUU ..................................................................................................((((((((.........))))))))....... (-10.70 = -10.46 + -0.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:38:04 2011