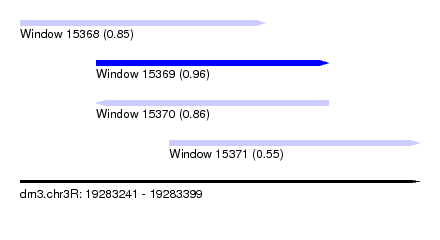
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,283,241 – 19,283,399 |
| Length | 158 |
| Max. P | 0.956042 |

| Location | 19,283,241 – 19,283,338 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 76.95 |
| Shannon entropy | 0.46080 |
| G+C content | 0.44494 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -16.28 |
| Energy contribution | -16.25 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.854540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
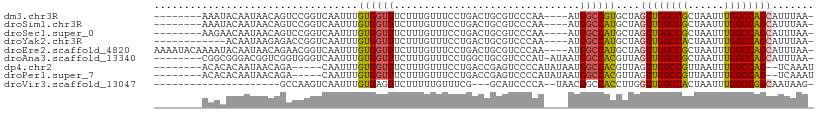
>dm3.chr3R 19283241 97 + 27905053 --------AAAUACAAUAACAGUCCGGUCAAUUUGUGGUUUCUUUGUUUCCUGACUGCGUCCCAA----AUGGCCGUGCUAGUUGGCCGCUAAUUUGGCCAGCAUUUAA- --------............((..((((((.((((.((.......(((....))).....)))))----)))))))..)).((((((((......))))))))......- ( -30.00, z-score = -2.64, R) >droSim1.chr3R 19103197 97 + 27517382 --------AAAUACAAUAACAGUCCGGUCAAUUUGUGGUUUCUUUGUUUCCUGACUGCGUCCCAA----AUGGCCAUGCUAGUUGGCCGCUAAUUUGGCCAGCAUUUAA- --------............(((..(((((.((((.((.......(((....))).....)))))----))))))..))).((((((((......))))))))......- ( -26.10, z-score = -1.45, R) >droSec1.super_0 19646405 97 + 21120651 --------AAGAACAAUAACAGUCCGGUCAAUUUGUGGUUUCUUUGUUUCCUGACUGCGUCCCAA----AUGGCCAUGCUAGUUGGCCGCUAAUUUGGCCAGCAUUUAA- --------............(((..(((((.((((.((.......(((....))).....)))))----))))))..))).((((((((......))))))))......- ( -26.10, z-score = -1.13, R) >droYak2.chr3R 20166401 93 + 28832112 ------------ACAAUAAGAGACCGGUCAAUUUGUGGUUUCUUUGUUUCCUGACUGCGUCCCAA----AUGGCCAUGCUAGUUGGCCACUAAUUUGGCCAGCAUUUAA- ------------.......(.(((((((((....(..(.....)..)....)))))).))).)..----.((((...))))((((((((......))))))))......- ( -26.30, z-score = -1.52, R) >droEre2.scaffold_4820 1703877 105 - 10470090 AAAAUACAAAAUACAAUAACAGAACGGUCAAUUUGUGGUUUCUUUGUUUCCUGACUGCGUCCCAA----AUGGCCAUGCUAGUUGGCCGCUAAUUUGGCCAGCAUUUAA- .........................(((((.((((.((.......(((....))).....)))))----))))))......((((((((......))))))))......- ( -25.10, z-score = -0.83, R) >droAna3.scaffold_13340 3426871 100 - 23697760 --------CGGCGGGACGGUCGGUGGGUCAAUUUGUGGUUUCUUUGUUUCCUGGCUGCGUCCCAU-AUAAUGGCCACGUUAGUUGGCCGCUAAUUUGGCCAGCAUUUAA- --------..(((((((((((((.(((.(((............))).))))))))..))))))..-....((((((.((((((.....)))))).))))))))......- ( -36.30, z-score = -1.76, R) >dp4.chr2 2766724 95 + 30794189 --------ACACACAAUAACAGA-----CAAUUUGUGGUUUCUUUGUUUCCUGACCGAGUCCCCAUAUAAUGGCCACGUUAGUUGGCCGUUAAUUUGGCCAG--UCAAAU --------.............((-----(....(((((...(((.(((....))).)))...)))))((((((((((....).))))))))).........)--)).... ( -24.30, z-score = -1.78, R) >droPer1.super_7 2859954 95 - 4445127 --------ACACACAAUAACAGA-----CAAUUUGUGGUUUCUUUGUUUCCUGACCGAGUCCCCAUAUAAUGGCCACGUUAGUUGGCCGUUAAUUUGGCCAG--UCAAAU --------.............((-----(....(((((...(((.(((....))).)))...)))))((((((((((....).))))))))).........)--)).... ( -24.30, z-score = -1.78, R) >droVir3.scaffold_13047 8915936 83 + 19223366 ---------------------GCCAAGUCAAUUUGUGAGUUCUUUUUGUUUCG---GCAUCCCCA--UAACGGCCACCUUGGUUGGCCACUAAUUUGGCCGGCAAUAAG- ---------------------(((..(((((.(((............(((..(---(.....)).--.)))((((((....).)))))..))).))))).)))......- ( -20.90, z-score = -0.52, R) >consensus ________AAACACAAUAACAGACCGGUCAAUUUGUGGUUUCUUUGUUUCCUGACUGCGUCCCAA____AUGGCCAUGCUAGUUGGCCGCUAAUUUGGCCAGCAUUUAA_ ..................................((((((............(((...)))..........))))))....((((((((......))))))))....... (-16.28 = -16.25 + -0.03)
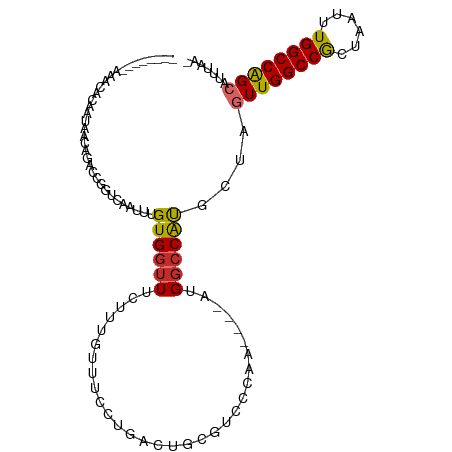
| Location | 19,283,271 – 19,283,363 |
|---|---|
| Length | 92 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 74.81 |
| Shannon entropy | 0.47928 |
| G+C content | 0.44115 |
| Mean single sequence MFE | -27.73 |
| Consensus MFE | -12.10 |
| Energy contribution | -11.74 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.956042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19283271 92 + 27905053 UUUCUUUGUUUCCUGACUGCGUCCC-A---AAUGGCCGUGCUAGUUGGCCGCUAAUUUGGCCAGCAUUUAAGCCGGUC--------------AGUCUUUAAUUAACAAAG- ...(((((((....((((((((...-.---.)))((((.(((.((((((((......)))))))).....))))))))--------------)))).......)))))))- ( -29.30, z-score = -3.20, R) >droSim1.chr3R 19103227 92 + 27517382 UUUCUUUGUUUCCUGACUGCGUCCC-A---AAUGGCCAUGCUAGUUGGCCGCUAAUUUGGCCAGCAUUUAAGCCGGUC--------------AGUCUUUAAUUAACAAAG- ...(((((((..(((((((.((...-.---..((((...))))((((((((......))))))))......)))))))--------------)).........)))))))- ( -27.90, z-score = -2.90, R) >droSec1.super_0 19646435 92 + 21120651 UUUCUUUGUUUCCUGACUGCGUCCC-A---AAUGGCCAUGCUAGUUGGCCGCUAAUUUGGCCAGCAUUUAAGCCGGUC--------------AGUCUUUAAUUAACAAAG- ...(((((((..(((((((.((...-.---..((((...))))((((((((......))))))))......)))))))--------------)).........)))))))- ( -27.90, z-score = -2.90, R) >droYak2.chr3R 20166427 92 + 28832112 UUUCUUUGUUUCCUGACUGCGUCCC-A---AAUGGCCAUGCUAGUUGGCCACUAAUUUGGCCAGCAUUUAAGCCGGUC--------------AGGCUUUAAUUAACAAAG- ...(((((((.((((((((.((...-.---..((((...))))((((((((......))))))))......)))))))--------------)))........)))))))- ( -32.10, z-score = -3.75, R) >droEre2.scaffold_4820 1703915 92 - 10470090 UUUCUUUGUUUCCUGACUGCGUCCC-A---AAUGGCCAUGCUAGUUGGCCGCUAAUUUGGCCAGCAUUUAAGCCGGUC--------------AGCCUUUAAUUAACAAAG- ...(((((((..(((((((.((...-.---..((((...))))((((((((......))))))))......)))))))--------------)).........)))))))- ( -27.90, z-score = -2.53, R) >droAna3.scaffold_13340 3426901 109 - 23697760 UUUCUUUGUUUCCUGGCUGCGUCCC-AUAUAAUGGCCACGUUAGUUGGCCGCUAAUUUGGCCAGCAUUUAAGGCGGCCUGUGUGUGGCUGCGAGGCUUUAAUUAACAAAG- ...(((((((....((((.(((.((-(((((..((((.(.(((((((((((......))))))))...))).).))))..)))))))..))).))))......)))))))- ( -43.80, z-score = -3.50, R) >dp4.chr2 2766749 96 + 30794189 UUUCUUUGUUUCCUGACCGAGUCCCCAUAUAAUGGCCACGUUAGUUGGCCGUUAAUUUGGCCAGU-CAAAUGACUGCG--------------AGGCUUUAAUUAAAAAAAG .......(((((....((((((.......((((((((((....).))))))))))))))).((((-(....))))).)--------------))))............... ( -24.81, z-score = -2.11, R) >droPer1.super_7 2859979 96 - 4445127 UUUCUUUGUUUCCUGACCGAGUCCCCAUAUAAUGGCCACGUUAGUUGGCCGUUAAUUUGGCCAGU-CAAAUGACUGCG--------------AGGCUUUAAUUAAAAAAAG .......(((((....((((((.......((((((((((....).))))))))))))))).((((-(....))))).)--------------))))............... ( -24.81, z-score = -2.11, R) >droVir3.scaffold_13047 8915953 92 + 19223366 GUUCUUUUUGUUUCGGCAUCCCCAUAAC-----GGCCACCUUGGUUGGCCACUAAUUUGGCCGGCAAUAAGACCACGCUAC-----------AUGGCUUAAUUAAAAA--- (.((((.((((..((((...........-----((((((....).))))).(......))))))))).)))).)..(((..-----------..)))...........--- ( -23.00, z-score = -0.69, R) >droGri2.scaffold_15074 7026267 96 - 7742996 GUUCUUUUUGUUUCGGCAUCUCAAUAAUAAUAUGACCACCUUAGUUG-CCACUAAUUUGGCCAGCAUUUAACCUGCGCCAC-----------AUGCCUCAAUUAAAAA--- ....((((.(((..(((((.....................(((((..-..)))))..(((((((........))).)))).-----------)))))..))).)))).--- ( -15.80, z-score = -0.93, R) >consensus UUUCUUUGUUUCCUGACUGCGUCCC_A___AAUGGCCACGCUAGUUGGCCGCUAAUUUGGCCAGCAUUUAAGCCGGUC______________AGGCUUUAAUUAACAAAG_ ...........................................((((((((......)))))))).............................................. (-12.10 = -11.74 + -0.36)
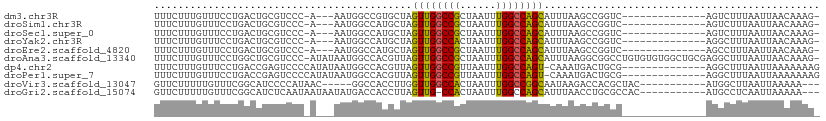
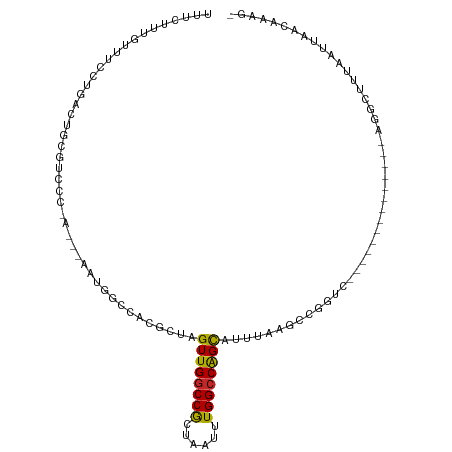
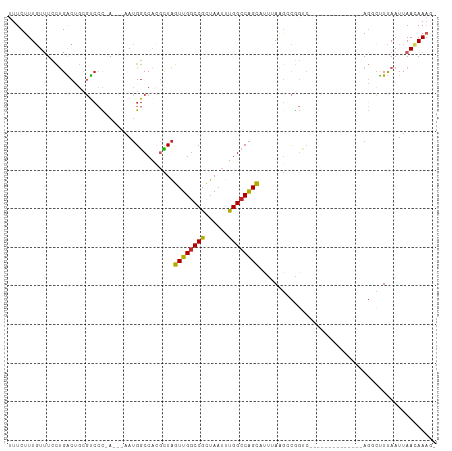
| Location | 19,283,271 – 19,283,363 |
|---|---|
| Length | 92 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 74.81 |
| Shannon entropy | 0.47928 |
| G+C content | 0.44115 |
| Mean single sequence MFE | -28.48 |
| Consensus MFE | -12.15 |
| Energy contribution | -12.67 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.857429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19283271 92 - 27905053 -CUUUGUUAAUUAAAGACU--------------GACCGGCUUAAAUGCUGGCCAAAUUAGCGGCCAACUAGCACGGCCAUU---U-GGGACGCAGUCAGGAAACAAAGAAA -(((((((..((...((((--------------(.(((((......)))))((((((....((((.........)))))))---)-))....)))))..)))))))))... ( -29.80, z-score = -2.91, R) >droSim1.chr3R 19103227 92 - 27517382 -CUUUGUUAAUUAAAGACU--------------GACCGGCUUAAAUGCUGGCCAAAUUAGCGGCCAACUAGCAUGGCCAUU---U-GGGACGCAGUCAGGAAACAAAGAAA -(((((((..((...((((--------------(.(((((......)))))((((((....(((((.......))))))))---)-))....)))))..)))))))))... ( -31.20, z-score = -3.31, R) >droSec1.super_0 19646435 92 - 21120651 -CUUUGUUAAUUAAAGACU--------------GACCGGCUUAAAUGCUGGCCAAAUUAGCGGCCAACUAGCAUGGCCAUU---U-GGGACGCAGUCAGGAAACAAAGAAA -(((((((..((...((((--------------(.(((((......)))))((((((....(((((.......))))))))---)-))....)))))..)))))))))... ( -31.20, z-score = -3.31, R) >droYak2.chr3R 20166427 92 - 28832112 -CUUUGUUAAUUAAAGCCU--------------GACCGGCUUAAAUGCUGGCCAAAUUAGUGGCCAACUAGCAUGGCCAUU---U-GGGACGCAGUCAGGAAACAAAGAAA -(((((((........(((--------------(((((((......)))).((((....(((((((.......))))))))---)-))......)))))).)))))))... ( -32.00, z-score = -2.95, R) >droEre2.scaffold_4820 1703915 92 + 10470090 -CUUUGUUAAUUAAAGGCU--------------GACCGGCUUAAAUGCUGGCCAAAUUAGCGGCCAACUAGCAUGGCCAUU---U-GGGACGCAGUCAGGAAACAAAGAAA -(((((((..((...((((--------------(.(((((......)))))((((((....(((((.......))))))))---)-))....)))))..)))))))))... ( -30.60, z-score = -2.41, R) >droAna3.scaffold_13340 3426901 109 + 23697760 -CUUUGUUAAUUAAAGCCUCGCAGCCACACACAGGCCGCCUUAAAUGCUGGCCAAAUUAGCGGCCAACUAACGUGGCCAUUAUAU-GGGACGCAGCCAGGAAACAAAGAAA -(((((((........(((.((.(((.......)))(((((...(((.((((((..((((.......))))..)))))).)))..-))).))..)).))).)))))))... ( -31.40, z-score = -1.04, R) >dp4.chr2 2766749 96 - 30794189 CUUUUUUUAAUUAAAGCCU--------------CGCAGUCAUUUG-ACUGGCCAAAUUAACGGCCAACUAACGUGGCCAUUAUAUGGGGACUCGGUCAGGAAACAAAGAAA ..((((((......(((((--------------(((((((....)-))))...........(((((.(....))))))......))))).))......(....))))))). ( -25.10, z-score = -1.84, R) >droPer1.super_7 2859979 96 + 4445127 CUUUUUUUAAUUAAAGCCU--------------CGCAGUCAUUUG-ACUGGCCAAAUUAACGGCCAACUAACGUGGCCAUUAUAUGGGGACUCGGUCAGGAAACAAAGAAA ..((((((......(((((--------------(((((((....)-))))...........(((((.(....))))))......))))).))......(....))))))). ( -25.10, z-score = -1.84, R) >droVir3.scaffold_13047 8915953 92 - 19223366 ---UUUUUAAUUAAGCCAU-----------GUAGCGUGGUCUUAUUGCCGGCCAAAUUAGUGGCCAACCAAGGUGGCC-----GUUAUGGGGAUGCCGAAACAAAAAGAAC ---........((((((((-----------(...))))).))))..((((((((......))))).((....))))).-----(((.(((.....))).)))......... ( -24.20, z-score = -0.19, R) >droGri2.scaffold_15074 7026267 96 + 7742996 ---UUUUUAAUUGAGGCAU-----------GUGGCGCAGGUUAAAUGCUGGCCAAAUUAGUGG-CAACUAAGGUGGUCAUAUUAUUAUUGAGAUGCCGAAACAAAAAGAAC ---(((((..((..(((((-----------.(.(.(((.......)))((((((..(((((..-..)))))..)))))).........).).)))))..))..)))))... ( -24.20, z-score = -1.67, R) >consensus _CUUUGUUAAUUAAAGCCU______________GACCGGCUUAAAUGCUGGCCAAAUUAGCGGCCAACUAACAUGGCCAUU___U_GGGACGCAGUCAGGAAACAAAGAAA .(((((((........................................((((((..((((.......))))..)))))).........((.....))....)))))))... (-12.15 = -12.67 + 0.52)
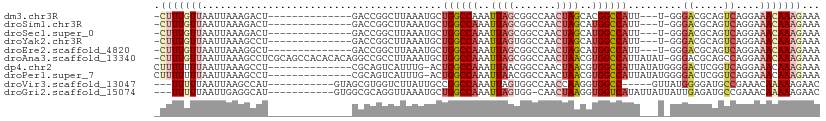

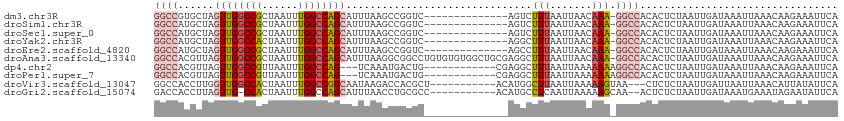
| Location | 19,283,300 – 19,283,399 |
|---|---|
| Length | 99 |
| Sequences | 10 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.00 |
| Shannon entropy | 0.37295 |
| G+C content | 0.39199 |
| Mean single sequence MFE | -24.70 |
| Consensus MFE | -13.69 |
| Energy contribution | -14.27 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.552382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19283300 99 + 27905053 GGCCGUGCUAGUUGGCCGCUAAUUUGGCCAGCAUUUAAGCCGGUC--------------AGUCUUUAAUUAACAAA-GGCCACACUCUAAUUGAUAAAUUAAACAAGAAAUUCA (((((.(((.((((((((......)))))))).....))))))))--------------.((((((.......)))-))).......(((((....)))))............. ( -26.80, z-score = -2.25, R) >droSim1.chr3R 19103256 99 + 27517382 GGCCAUGCUAGUUGGCCGCUAAUUUGGCCAGCAUUUAAGCCGGUC--------------AGUCUUUAAUUAACAAA-GGCCACACUCUAAUUGAUAAAUUAAACAAGAAAUUCA ((((..(((.((((((((......)))))))).....))).))))--------------.((((((.......)))-))).......(((((....)))))............. ( -22.70, z-score = -1.13, R) >droSec1.super_0 19646464 99 + 21120651 GGCCAUGCUAGUUGGCCGCUAAUUUGGCCAGCAUUUAAGCCGGUC--------------AGUCUUUAAUUAACAAA-GGCCACACUCUAAUUGAUAAAUUAAACAAGAAAUUCA ((((..(((.((((((((......)))))))).....))).))))--------------.((((((.......)))-))).......(((((....)))))............. ( -22.70, z-score = -1.13, R) >droYak2.chr3R 20166456 99 + 28832112 GGCCAUGCUAGUUGGCCACUAAUUUGGCCAGCAUUUAAGCCGGUC--------------AGGCUUUAAUUAACAAA-GGCCACACUCUAAUUGAUAAAUUAAACAAGAAAUUCA ((((......((((((((......))))))))(((.(((((....--------------.))))).))).......-))))......(((((....)))))............. ( -29.80, z-score = -3.40, R) >droEre2.scaffold_4820 1703944 99 - 10470090 GGCCAUGCUAGUUGGCCGCUAAUUUGGCCAGCAUUUAAGCCGGUC--------------AGCCUUUAAUUAACAAA-GGCCACACUCUAAUUGAUAAAUUAAACAAGAAAUUCA ((((..(((.((((((((......)))))))).....))).))))--------------.((((((.......)))-))).......(((((....)))))............. ( -25.40, z-score = -1.72, R) >droAna3.scaffold_13340 3426933 113 - 23697760 GGCCACGUUAGUUGGCCGCUAAUUUGGCCAGCAUUUAAGGCGGCCUGUGUGUGGCUGCGAGGCUUUAAUUAACAAA-GGCCACACUCUAAUUGAUAAAUUAAACAAGAAAUUCA ((((.(.(((((((((((......))))))))...))).).))))(((((((((((....................-))))))))..(((((....))))).)))......... ( -35.65, z-score = -1.70, R) >dp4.chr2 2766782 99 + 30794189 GGCCACGUUAGUUGGCCGUUAAUUUGGCCAG---UCAAAUGACUG------------CGAGGCUUUAAUUAAAAAAAGGCCACACUCUAAUUGAUAAAUUAAACAAGAAAUUCA ((((((....).))))).((((((((((.((---((....)))))------------)..((((((.........)))))).............))))))))............ ( -24.90, z-score = -2.25, R) >droPer1.super_7 2860012 99 - 4445127 GGCCACGUUAGUUGGCCGUUAAUUUGGCCAG---UCAAAUGACUG------------CGAGGCUUUAAUUAAAAAAAGGCCACACUCUAAUUGAUAAAUUAAACAAGAAAUUCA ((((((....).))))).((((((((((.((---((....)))))------------)..((((((.........)))))).............))))))))............ ( -24.90, z-score = -2.25, R) >droVir3.scaffold_13047 8915981 100 + 19223366 GGCCACCUUGGUUGGCCACUAAUUUGGCCGGCAAUAAGACCACGCU-----------ACAUGGCUUAAUUAAAAAGUAA---CUCUCUAAUUGAUUAAUUAAACAUUAUAUUCA .((((.....((((((((......))))))))....((......))-----------...))))((((((((..((...---....))..))))))))................ ( -19.70, z-score = -0.92, R) >droGri2.scaffold_15074 7026300 100 - 7742996 GACCACCUUAGUUG-CCACUAAUUUGGCCAGCAUUUAACCUGCGCC-----------ACAUGCCUCAAUUAAAAAGCAA--ACUCUCUAAUUGAUAAAUGAAAUAGAAUAUUCA .......(((((..-..)))))..(((((((........))).)))-----------)(((...(((((((.(.((...--.)).).)))))))...))).............. ( -14.40, z-score = -0.89, R) >consensus GGCCACGCUAGUUGGCCGCUAAUUUGGCCAGCAUUUAAGCCGGUC______________AGGCUUUAAUUAACAAA_GGCCACACUCUAAUUGAUAAAUUAAACAAGAAAUUCA ((((......((((((((......))))))))...............................(((.......))).))))................................. (-13.69 = -14.27 + 0.58)
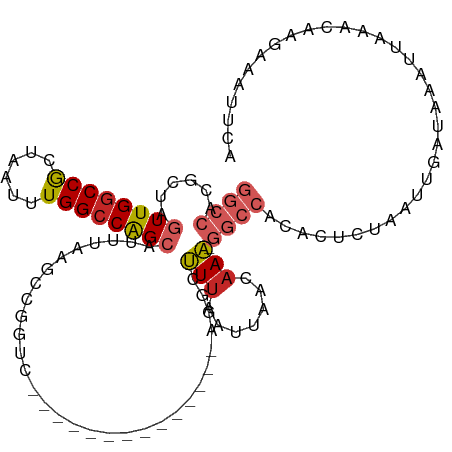
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:38:02 2011