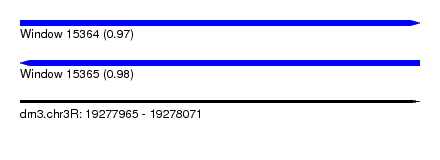
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,277,965 – 19,278,071 |
| Length | 106 |
| Max. P | 0.976989 |

| Location | 19,277,965 – 19,278,071 |
|---|---|
| Length | 106 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 80.22 |
| Shannon entropy | 0.38538 |
| G+C content | 0.31899 |
| Mean single sequence MFE | -26.08 |
| Consensus MFE | -15.07 |
| Energy contribution | -15.58 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.966742 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
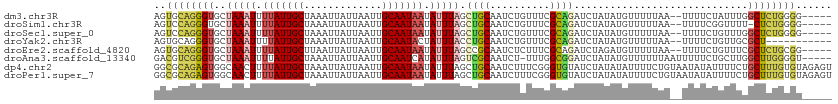
>dm3.chr3R 19277965 106 + 27905053 AGUGCAGGGUGCUAAAUUUUAUUGCUAAAUUAUUAAUUGCAAUAAUAUUUAGCUGCAAUCUGUUUCGCAGAUCUAUAUGUUUUUAA--UUUUCUAUUUGGCUCUGGGG----- ....(((((((((((((.(((((((.............))))))).)))))))....(((((.....)))))..............--...........))))))...----- ( -26.32, z-score = -2.43, R) >droSim1.chr3R 19097924 105 + 27517382 AGUCCAGGGUGCUAAAUUUUAUUGCUAAAUUAUUAAUUGCAAUAAUAUUUAGCUGCAAUCUGUUUCGCAGAUCUAUAUGUUUUUAA--UUUUCGGUUUU-CUCUGGGG----- ..(((((((.(((((((.(((((((.............))))))).)))))))....(((((.....)))))..............--...........-))))))).----- ( -27.72, z-score = -3.85, R) >droSec1.super_0 19640742 106 + 21120651 AGUCCAGGGUGCUAAAUUUUAUUGCUAAAUUAUUAAUUGCAAUAAUAUUUAGCUGCAAUCUGUUUCGCAGAUCUAUAUGUUUUUAA--UUUUCUGUUUGGCUCUGGGG----- ..(((((((((((((((.(((((((.............))))))).)))))))....(((((.....)))))..............--...........)))))))).----- ( -30.12, z-score = -3.93, R) >droYak2.chr3R 20160828 100 + 28832112 AGUGCAGGGUGCUAAAUUUUAUUGCUAAAUUAUUAAUUGCAAUACUAUUUACCUGCAAUCUGUUUCGCAGAUCUAUAUGUUUUUAA--UUUUCUGUUGCGCU----------- (((((((.(((((((((..((((((.............))))))..)))))...)))(((((.....)))))..............--.....).)))))))----------- ( -20.72, z-score = -1.24, R) >droEre2.scaffold_4820 1698643 106 - 10470090 AGUGCAGGGUGCUAAAUUUUAUUGCUUAAUUAUUAAUUGCAAUAAUAUUUAGCCGCAAUCUCUUUCGCAGAUCUAGAUGUUUUUAA--UUUUCUGUUUCGCUCUGCGG----- ..(((((((((((((((.(((((((.............))))))).))))))).............(((((..((((....)))).--...)))))...)))))))).----- ( -29.42, z-score = -3.54, R) >droAna3.scaffold_13340 3421490 107 - 23697760 GACGUCGGGUGCUAAAUUUUAUUGCUAAAUUAUUAAUUGCAAUCAUAUUUAGUCGCAAUCU-UUUGGCGGAUCUAUAUGUUUUUUAAUUUUUCUGCUUGGCUUGGGGU----- .((.(((((((((((((...(((((.............)))))...)))))))........-...((((((....................))))))..)))))).))----- ( -20.77, z-score = -0.95, R) >dp4.chr2 2761329 113 + 30794189 GGCGCAGAGUGGCAACUUUUAUUGCUAAAUUAUUAAUUGCAAUAAUAUUUAGCUGCAAUCUUUCGGGUGUAUCUAUAUAUUUUCUGUAAUAUAUUUUCUGCUUUGUGUAGAGU .((((((((((((((......))))).........((((((..(((((.(((.((((..(....)..)))).))).)))))...)))))).........)))))))))..... ( -26.80, z-score = -1.80, R) >droPer1.super_7 2854586 113 - 4445127 GGCGCAGAGUGGCAACUUUUAUUGCUAAAUUAUUAAUUGCAAUAAUAUUUAGCUGCAAUCUUUCGGGUGUAUCUAUAUAUUUUCUGUAAUAUAUUUUCUGCUUUGUGUAGAGU .((((((((((((((......))))).........((((((..(((((.(((.((((..(....)..)))).))).)))))...)))))).........)))))))))..... ( -26.80, z-score = -1.80, R) >consensus AGUGCAGGGUGCUAAAUUUUAUUGCUAAAUUAUUAAUUGCAAUAAUAUUUAGCUGCAAUCUGUUUCGCAGAUCUAUAUGUUUUUAA__UUUUCUGUUUGGCUCUGGGG_____ ..((((((((..(((((.(((((((.............))))))).))))).((((..........)))).............................))))))))...... (-15.07 = -15.58 + 0.52)
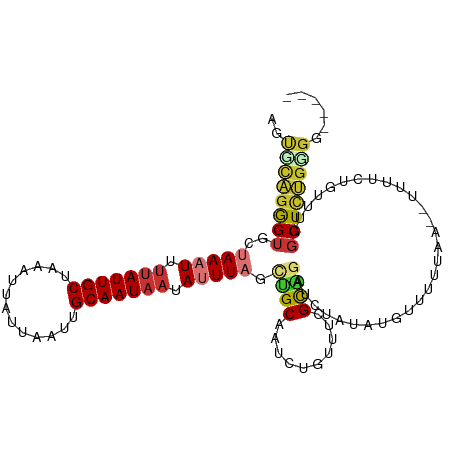
| Location | 19,277,965 – 19,278,071 |
|---|---|
| Length | 106 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 80.22 |
| Shannon entropy | 0.38538 |
| G+C content | 0.31899 |
| Mean single sequence MFE | -21.27 |
| Consensus MFE | -7.77 |
| Energy contribution | -8.38 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.48 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.976989 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 19277965 106 - 27905053 -----CCCCAGAGCCAAAUAGAAAA--UUAAAAACAUAUAGAUCUGCGAAACAGAUUGCAGCUAAAUAUUAUUGCAAUUAAUAAUUUAGCAAUAAAAUUUAGCACCCUGCACU -----...(((..............--.............((((((.....))))))...(((((((.(((((((.............))))))).)))))))...))).... ( -21.02, z-score = -3.77, R) >droSim1.chr3R 19097924 105 - 27517382 -----CCCCAGAG-AAAACCGAAAA--UUAAAAACAUAUAGAUCUGCGAAACAGAUUGCAGCUAAAUAUUAUUGCAAUUAAUAAUUUAGCAAUAAAAUUUAGCACCCUGGACU -----..((((..-...........--.............((((((.....))))))...(((((((.(((((((.............))))))).)))))))...))))... ( -25.02, z-score = -5.89, R) >droSec1.super_0 19640742 106 - 21120651 -----CCCCAGAGCCAAACAGAAAA--UUAAAAACAUAUAGAUCUGCGAAACAGAUUGCAGCUAAAUAUUAUUGCAAUUAAUAAUUUAGCAAUAAAAUUUAGCACCCUGGACU -----..((((..............--.............((((((.....))))))...(((((((.(((((((.............))))))).)))))))...))))... ( -25.02, z-score = -5.23, R) >droYak2.chr3R 20160828 100 - 28832112 -----------AGCGCAACAGAAAA--UUAAAAACAUAUAGAUCUGCGAAACAGAUUGCAGGUAAAUAGUAUUGCAAUUAAUAAUUUAGCAAUAAAAUUUAGCACCCUGCACU -----------..............--.............((((((.....))))))((((((((((..((((((.............))))))..)))))....)))))... ( -19.72, z-score = -2.15, R) >droEre2.scaffold_4820 1698643 106 + 10470090 -----CCGCAGAGCGAAACAGAAAA--UUAAAAACAUCUAGAUCUGCGAAAGAGAUUGCGGCUAAAUAUUAUUGCAAUUAAUAAUUAAGCAAUAAAAUUUAGCACCCUGCACU -----..((((.((.....(((...--.........))).(((((.(....)))))))).(((((((.(((((((.............))))))).)))))))...))))... ( -27.12, z-score = -4.25, R) >droAna3.scaffold_13340 3421490 107 + 23697760 -----ACCCCAAGCCAAGCAGAAAAAUUAAAAAACAUAUAGAUCCGCCAAA-AGAUUGCGACUAAAUAUGAUUGCAAUUAAUAAUUUAGCAAUAAAAUUUAGCACCCGACGUC -----............((................................-.(((((((((.......).)))))))).....((((....)))).....)).......... ( -9.80, z-score = -0.48, R) >dp4.chr2 2761329 113 - 30794189 ACUCUACACAAAGCAGAAAAUAUAUUACAGAAAAUAUAUAGAUACACCCGAAAGAUUGCAGCUAAAUAUUAUUGCAAUUAAUAAUUUAGCAAUAAAAGUUGCCACUCUGCGCC ............(((((..(((((((......))))))).........(....)...((((((.....(((((((.............))))))).))))))...)))))... ( -21.22, z-score = -3.04, R) >droPer1.super_7 2854586 113 + 4445127 ACUCUACACAAAGCAGAAAAUAUAUUACAGAAAAUAUAUAGAUACACCCGAAAGAUUGCAGCUAAAUAUUAUUGCAAUUAAUAAUUUAGCAAUAAAAGUUGCCACUCUGCGCC ............(((((..(((((((......))))))).........(....)...((((((.....(((((((.............))))))).))))))...)))))... ( -21.22, z-score = -3.04, R) >consensus _____CCCCAGAGCAAAACAGAAAA__UUAAAAACAUAUAGAUCUGCGAAACAGAUUGCAGCUAAAUAUUAUUGCAAUUAAUAAUUUAGCAAUAAAAUUUAGCACCCUGCACU ........................................................(((((.(((((.(((((((.............))))))).))))).....))))).. ( -7.77 = -8.38 + 0.61)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:37:58 2011