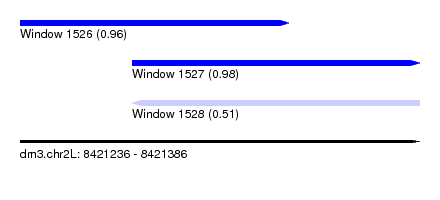
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,421,236 – 8,421,386 |
| Length | 150 |
| Max. P | 0.984219 |

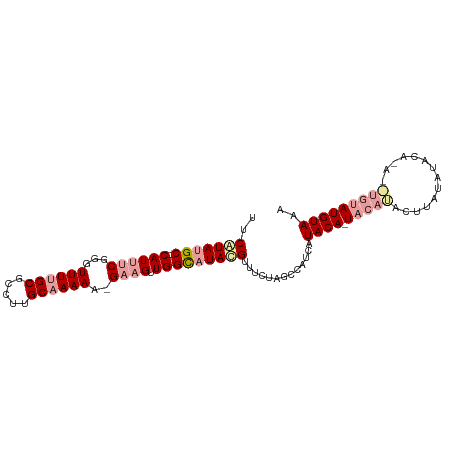
| Location | 8,421,236 – 8,421,337 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 79.93 |
| Shannon entropy | 0.28098 |
| G+C content | 0.35745 |
| Mean single sequence MFE | -26.67 |
| Consensus MFE | -11.21 |
| Energy contribution | -13.44 |
| Covariance contribution | 2.23 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.54 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8421236 101 + 23011544 UUCGUAUGCGCCAUUUCGGGUUUUGCGCCUUGCAAAAAAGAAGUUGGCAUACGUUUCUAGCCAUCAUACAAUUCAUACAUAUAUACAUACAUAUAUGUAAA ..(((((((.(.(((((...((((((.....))))))..))))).))))))))......................(((((((((......))))))))).. ( -26.80, z-score = -3.35, R) >droEre2.scaffold_4929 17329950 88 + 26641161 UUCAUAUGC--CAUUUCGGGUUUUGCGCCUUGCAAAA--GUUGUUGGUAUAUGUUUCUAGCCAUCAUACA-UACAUACUUAUAU--------GUAUGUAAA ..(((((((--((...(((.((((((.....))))))--.))).))))))))).............((((-((((((....)))--------))))))).. ( -29.00, z-score = -4.75, R) >droYak2.chr2L 11059084 96 + 22324452 UUCUUAGGC--CAUUUCGGGUUU-GCGCAUUGCAAAAA-GAAGUUGGCAUACGCUUCUAGCCAUCAUACA-UACAAACUAACAUACAGAUUUGUAUGUAAA .......((--((((((...(((-((.....)))))..-)))).))))....((.....)).....((((-((((((((.......)).)))))))))).. ( -24.20, z-score = -2.52, R) >consensus UUCAUAUGC__CAUUUCGGGUUUUGCGCCUUGCAAAAA_GAAGUUGGCAUACGUUUCUAGCCAUCAUACA_UACAUACUUAUAUACA_A__UGUAUGUAAA ..(((((((...(((((...((((((.....))))))..)))))..)))))))..................(((((((..............))))))).. (-11.21 = -13.44 + 2.23)

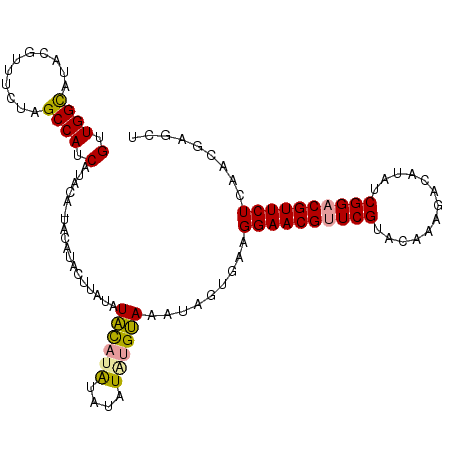
| Location | 8,421,278 – 8,421,386 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 84.21 |
| Shannon entropy | 0.21874 |
| G+C content | 0.38005 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -16.13 |
| Energy contribution | -15.92 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.71 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.984219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8421278 108 + 23011544 GUUGGCAUACGUUUCUAGCCAUCAUACAAUUCAUACAUAUAUACAUACAUAUAUGUAAAUAGUGAAGGAACGGUCGUACAAAGACAUAUCGGACGUUCUCAACGAGCU (.((((...........)))).)......(((.(((((((((......)))))))))....((...((((((..((.............))..))))))..))))).. ( -22.22, z-score = -1.55, R) >droEre2.scaffold_4929 17329988 106 + 26641161 GUUGGUAUAUGUUUCUAGCCAUCAUACA-UACAUACUUAUAUGUAUGUAAAUG-GCAGAAUAUGAAGGAACGUUCGUACAAAGACAUAUCGGACGUUCUCAACGAGCU ((((...((((((.((.(((((..((((-((((((....)))))))))).)))-))))))))))..((((((((((.............))))))))))))))..... ( -36.22, z-score = -5.19, R) >droYak2.chr2L 11059122 107 + 22324452 GUUGGCAUACGCUUCUAGCCAUCAUACA-UACAAACUAACAUACAGAUUUGUAUGUAAAUGGUGAAGGAACGUUCGUACAAAGACAUAUCGGACGUUCUCAACGAGCU (((((......((((..(((((..((((-((((((((.......)).)))))))))).)))))))))(((((((((.............))))))))))))))..... ( -33.12, z-score = -4.39, R) >consensus GUUGGCAUACGUUUCUAGCCAUCAUACA_UACAUACUUAUAUACAUAUAUAUAUGUAAAUAGUGAAGGAACGUUCGUACAAAGACAUAUCGGACGUUCUCAACGAGCU (.((((...........)))).)..................((((((....)))))).........((((((((((.............))))))))))......... (-16.13 = -15.92 + -0.21)

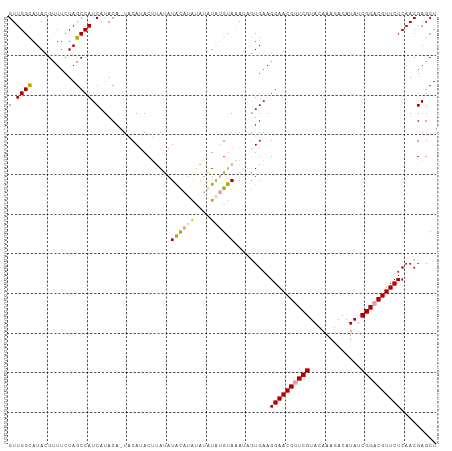
| Location | 8,421,278 – 8,421,386 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 84.21 |
| Shannon entropy | 0.21874 |
| G+C content | 0.38005 |
| Mean single sequence MFE | -28.57 |
| Consensus MFE | -14.94 |
| Energy contribution | -15.40 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.509785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8421278 108 - 23011544 AGCUCGUUGAGAACGUCCGAUAUGUCUUUGUACGACCGUUCCUUCACUAUUUACAUAUAUGUAUGUAUAUAUGUAUGAAUUGUAUGAUGGCUAGAAACGUAUGCCAAC ...((((((((((((..(((((......))).))..)))))..))).......((((((((((....))))))))))......))))((((...........)))).. ( -21.80, z-score = -0.12, R) >droEre2.scaffold_4929 17329988 106 - 26641161 AGCUCGUUGAGAACGUCCGAUAUGUCUUUGUACGAACGUUCCUUCAUAUUCUGC-CAUUUACAUACAUAUAAGUAUGUA-UGUAUGAUGGCUAGAAACAUAUACCAAC .....((((.((((((.(((((......))).)).)))))).......((((((-((((((((((((((....))))))-)))).)))))).))))........)))) ( -33.40, z-score = -4.78, R) >droYak2.chr2L 11059122 107 - 22324452 AGCUCGUUGAGAACGUCCGAUAUGUCUUUGUACGAACGUUCCUUCACCAUUUACAUACAAAUCUGUAUGUUAGUUUGUA-UGUAUGAUGGCUAGAAGCGUAUGCCAAC .((.((((..((((((.(((((......))).)).))))))((...(((((((((((((((.(((.....)))))))))-)))).)))))..)).))))...)).... ( -30.50, z-score = -2.85, R) >consensus AGCUCGUUGAGAACGUCCGAUAUGUCUUUGUACGAACGUUCCUUCACAAUUUACAUAUAUAUAUGUAUAUAAGUAUGUA_UGUAUGAUGGCUAGAAACGUAUGCCAAC .....((((.((((((.(((((......))).)).))))))((...((((((((((((((((..........))))))).)))).)))))..))..........)))) (-14.94 = -15.40 + 0.46)

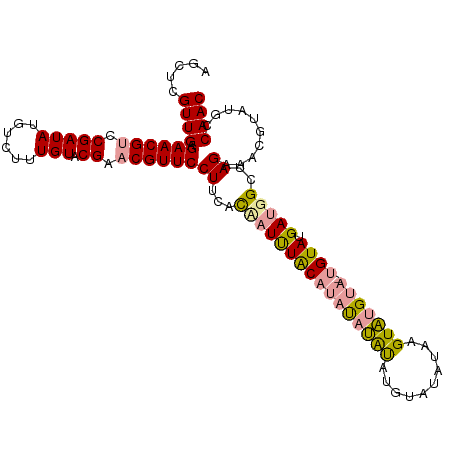
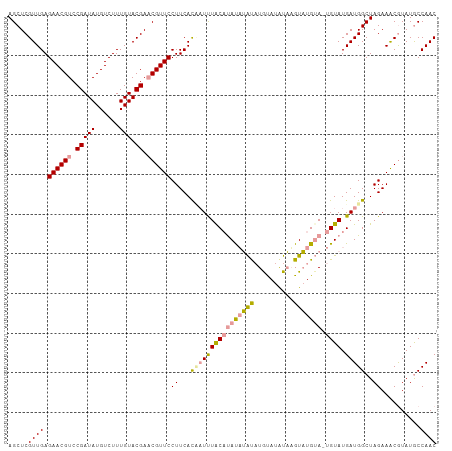
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:25:56 2011