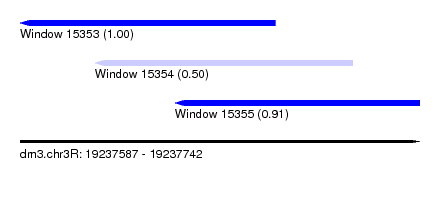
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,237,587 – 19,237,742 |
| Length | 155 |
| Max. P | 0.995979 |

| Location | 19,237,587 – 19,237,686 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 67.31 |
| Shannon entropy | 0.72070 |
| G+C content | 0.60318 |
| Mean single sequence MFE | -31.93 |
| Consensus MFE | -17.85 |
| Energy contribution | -17.60 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.78 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.995979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
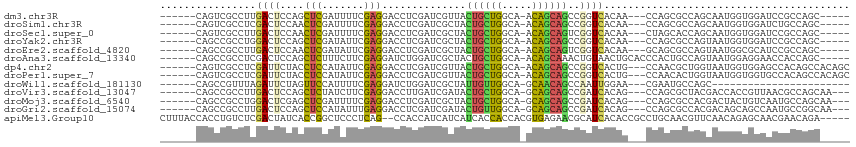
>dm3.chr3R 19237587 99 - 27905053 GGACCUCGAUCGUUACUGCUGGCAACAGCAGCCGGUCACAACCAGCGC--CAGCAAUGGUGGAU--CCGC-----CAG---CACCAAUCAGCCCAGUCCCACCUCGCCGCA ((((...(((((...((((((....)))))).))))).......((..--..))..(((((...--.)))-----)).---..............))))............ ( -31.90, z-score = -1.16, R) >droSim1.chr3R 19055797 99 - 27517382 GGACCUCGAUCGCUACUGCUGGCAACAGCAGCCGGUCACAACCAGCGC--CAGCAAUGGUGGAU--CUGC-----CAG---CACCAAUCAGACCAGUCCCACCUCGCCGCA ((.....(((((...((((((....)))))).)))))....)).(((.--..((...(((((..--(((.-----...---(........)..)))..)))))..))))). ( -33.40, z-score = -1.34, R) >droSec1.super_0 19600557 99 - 21120651 GGACCUCGAUCGCUACUGCUGGCAACAGCAGUCGGUCACAACUAGCAC--CAGCAAUGGUGGAU--CCGC-----CAG---CACCAAUCAGCCCAGUCCCACCUCGCCGCA ((((...(((((..(((((((....))))))))))))...........--..((..(((((...--.)))-----)))---).............))))............ ( -33.40, z-score = -2.07, R) >droYak2.chr3R 20117590 99 - 28832112 GGACCUCGAUCGCUACUGCUGGCAACAGCAGCCGGUCACAACCAGCGC--CAGUAAUGGUGGAU--CCGC-----CAG---CACCAAUCAGCCCAGUCCCACCUCGCCGCA ((((...(((((...((((((....)))))).))))).......((..--..((..(((((...--.)))-----)).---.))......))...))))............ ( -32.50, z-score = -1.30, R) >droEre2.scaffold_4820 1657793 99 + 10470090 GGACCUCGAUCGCUACUGCUGGCAACAGCAGUCGGUCACAAGCAGCGC--CAGUAAUGGCGCAU--CCGC-----CAG---CACCAAUCAGCCCAGUCCCACCUCGCCGCA ((((...(((((..(((((((....))))))))))))....((.((((--((....))))))..--..(.-----...---)........))...))))............ ( -38.80, z-score = -3.72, R) >dp4.chr2 2719226 105 - 30794189 GGACCUCGAUCGUUACUGCUGGCAACAGCAGCCGGUCACUGCCAACGC--UGGUAAUGGUGGAG--CCACAGCCACAG--CCACCAACCAGCCCAGUCCCACGUCUCCUCA ((((...(((((...((((((....)))))).))))).........((--((((..((((((.(--(....)).....--))))))))))))...))))............ ( -42.50, z-score = -3.29, R) >droPer1.super_7 2812414 105 + 4445127 GGACCUCGAUCGUUACUGCUGGCAACAGCAGCCGGUCACUGCCAACAC--UGGUAAUGGUGGUG--CCACAGCCACAG--CCACCAACCAGCCCAGUCCCACGUCUCCUCA ((((...(((((...((((((....)))))).)))))..........(--((((..((((((((--(....))....)--)))))))))))....))))............ ( -40.20, z-score = -2.73, R) >droWil1.scaffold_181130 9272945 78 - 16660200 GGAUCUGGAUCGCUAUUGUUGGCAGCAACAGCCAAUUGGAACGAAUGC---------------------------------CAGCACUCAGCCAAGUCGUACAUCACCGCA .((..(((........((((((((.......((....))......)))---------------------------------))))).....)))..))............. ( -19.24, z-score = 0.16, R) >droVir3.scaffold_13047 8871199 111 - 19223366 GGACCUUGAUCGAUACUGCUGGCAGCAGCAGCCGAUCACAGCCAGCGCUACGACCACCGUUAACGCCAGCAAUGUGCGAUGUACUAACCAGCCAAGUCCCACAUCUCCGCA (((...((((((...((((((....)))))).))))))..((..(((..(((.....)))...)))..)).(((((.(((...((....))....))).))))).)))... ( -31.40, z-score = -1.21, R) >droMoj3.scaffold_6540 2348628 111 + 34148556 GGACCUCGAUCGCUACUGCUGGCAGCAGCAGCCGAUCACAGCCAGCGCCACGACUACUGUCAAUGCCAGCAACGUGCGCUGCAGCAACCAGCCAAGCCCCACGUCUCCGCA (((....(((((...((((((....)))))).)))))...((((((((.(((((....)))..((....))..))))))))..))....................)))... ( -34.40, z-score = 0.05, R) >droGri2.scaffold_15074 6980784 111 + 7742996 GGACCUCGAUCGAUACUGUUGGCAGCAGCAGCCGAUCACAGCCAGCGCCACGACAGCAGCCAAUGCCGGCAAUGUUCGAUGUAGCAACCAACCCAGUCCCACAUCACCACA ((((...(((((...((((((....)))))).))))).......((..(((((((...(((......)))..)).))).))..))..........))))............ ( -29.70, z-score = -1.12, R) >apiMel3.Group10 4074115 98 + 11440700 CCAUCAUCAUCACCACCACGUGAGAACGCAUCACACCGCCUGCAACGU--UCAACAGAGCAACGAACAG-----------ACACCGAUGGACCAAUCCGACUGCGACUAUU ......((.((((......)))))).((((((.....(((((......--....))).)).........-----------........(((....))))).))))...... ( -15.70, z-score = -0.36, R) >consensus GGACCUCGAUCGCUACUGCUGGCAACAGCAGCCGGUCACAACCAGCGC__CAGCAAUGGUGGAU__CCGC_____CAG___CACCAAUCAGCCCAGUCCCACCUCGCCGCA ((((...(((((...((((((....)))))).))))).....................(((......))).........................))))............ (-17.85 = -17.60 + -0.25)
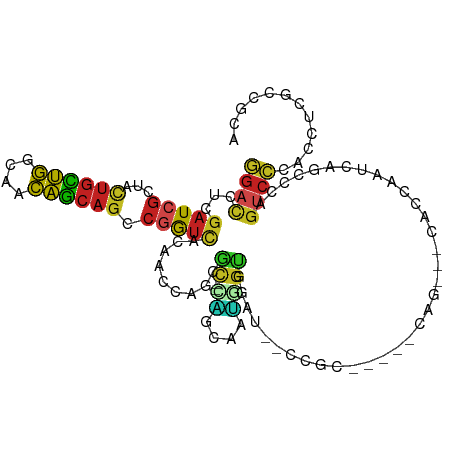
| Location | 19,237,616 – 19,237,716 |
|---|---|
| Length | 100 |
| Sequences | 13 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 69.58 |
| Shannon entropy | 0.64234 |
| G+C content | 0.57858 |
| Mean single sequence MFE | -32.92 |
| Consensus MFE | -10.02 |
| Energy contribution | -9.80 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.92 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
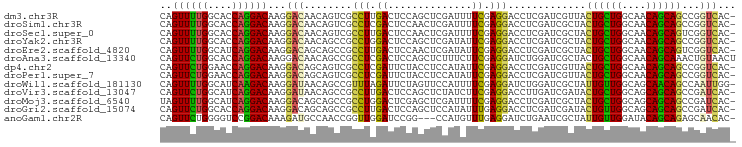
>dm3.chr3R 19237616 100 - 27905053 ------CAGUCGCCUUGACUCCAGCUCGAUUUUCGAGGACCUCGAUCGUUACUGCUGGCA-ACAGCAGCCGGUCACAA---CCAGCGCCAGCAAUGGUGGAUCCGCCAGC----- ------.((((.....))))...(((((.....)))(((((..(((((...((((((...-.)))))).)))))....---.....((((....)))))).)))))....----- ( -34.50, z-score = -1.26, R) >droSim1.chr3R 19055826 100 - 27517382 ------CAGUCGCCUCGACUCCAACUCGAUUUUCGAGGACCUCGAUCGCUACUGCUGGCA-ACAGCAGCCGGUCACAA---CCAGCGCCAGCAAUGGUGGAUCUGCCAGC----- ------.(((((...)))))....((((.....))))......(((((((.((((((...-.))))))..(((....)---)))))((((....)))).)))).......----- ( -35.90, z-score = -1.85, R) >droSec1.super_0 19600586 100 - 21120651 ------CAGUCGCCUUGACUCCAACUCGAUUUUCGAGGACCUCGAUCGCUACUGCUGGCA-ACAGCAGUCGGUCACAA---CUAGCACCAGCAAUGGUGGAUCCGCCAGC----- ------.((((.....))))....((((.....))))......(((((..(((((((...-.))))))))))))....---...((....))..(((((....)))))..----- ( -35.40, z-score = -2.37, R) >droYak2.chr3R 20117619 100 - 28832112 ------CAGCCGCCUGGACUCCAGCUCGAUAUUCGAGGACCUCGAUCGCUACUGCUGGCA-ACAGCAGCCGGUCACAA---CCAGCGCCAGUAAUGGUGGAUCCGCCAGC----- ------.....((((((.......((((.....))))......(((((...((((((...-.)))))).)))))....---)))).))......(((((....)))))..----- ( -38.70, z-score = -1.94, R) >droEre2.scaffold_4820 1657822 100 + 10470090 ------CAGCCGCCUUGACUCCAACUCGAUAUUCGAGGACCUCGAUCGCUACUGCUGGCA-ACAGCAGUCGGUCACAA---GCAGCGCCAGUAAUGGCGCAUCCGCCAGC----- ------..((.((.(((.......((((.....))))......(((((..(((((((...-.)))))))))))).)))---)).((((((....))))))....))....----- ( -43.00, z-score = -4.26, R) >droAna3.scaffold_13340 3380543 103 + 23697760 ------CAGCCGCCUCGACUCCAGCUCUUUCUUCGAGGAUCUGGAUCGCUACUGCUGGCA-ACAGCAAACUGUAACUGCACCCACUGCCAGUAAUGGAGGAACCACCAGC----- ------......((((((.(((((.(((((....))))).))))))).....((((((((-((((....))))............))))))))...))))..........----- ( -30.30, z-score = -0.92, R) >dp4.chr2 2719256 105 - 30794189 ------CAGUCGCCUCGAUUCUACCUCCAUAUUCGAGGACCUCGAUCGUUACUGCUGGCA-ACAGCAGCCGGUCACUG---CCAACGCUGGUAAUGGUGGAGCCACAGCCACAGC ------((((..((((((..............)))))).....(((((...((((((...-.)))))).)))))))))---.....((((......(((....))).....)))) ( -36.94, z-score = -1.86, R) >droPer1.super_7 2812444 105 + 4445127 ------CAGUCGCCUCGAUUCUACCUCCAUAUUCGAGGACCUCGAUCGUUACUGCUGGCA-ACAGCAGCCGGUCACUG---CCAACACUGGUAAUGGUGGUGCCACAGCCACAGC ------..(..(((((((..............)))))).)..)(((((...((((((...-.)))))).)))))..((---(((....)))))...(((((......)))))... ( -37.94, z-score = -2.38, R) >droWil1.scaffold_181130 9272971 82 - 16660200 ------CAGCCGUUUAGAUUCUAGUUCCAUUUUCGAGGAUCUGGAUCGCUAUUGUUGGCA-GCAACAGCCAAUUGGAA---CGAAUGCCAGC----------------------- ------.(((.(((((((((((.(.........).))))))))))).)))...(((((((-.......((....))..---....)))))))----------------------- ( -25.32, z-score = -2.16, R) >droVir3.scaffold_13047 8871238 102 - 19223366 ------CAGCCGCCUUGACUCCAGCUCUAUCUUCGAGGACCUUGAUCGAUACUGCUGGCA-GCAGCAGCCGAUCACAG---CCAGCGCUACGACCACCGUUAACGCCAGCAA--- ------......((((((..............))))))....((((((...((((((...-.)))))).))))))..(---(..(((..(((.....)))...)))..))..--- ( -28.84, z-score = -0.85, R) >droMoj3.scaffold_6540 2348667 102 + 34148556 ------CAGCCGCCUGGACUCGAGCUCGAUUUUCGAGGACCUCGAUCGCUACUGCUGGCA-GCAGCAGCCGAUCACAG---CCAGCGCCACGACUACUGUCAAUGCCAGCAA--- ------..((.((((((.((.(((((((.....))))...)))(((((...((((((...-.)))))).)))))..))---)))).))...(((....))).......))..--- ( -35.10, z-score = -1.02, R) >droGri2.scaffold_15074 6980823 102 + 7742996 ------CAGCCGCCUUGACUCCAGCUCCAUAUUUGAGGACCUCGAUCGAUACUGUUGGCA-GCAGCAGCCGAUCACAG---CCAGCGCCACGACAGCAGCCAAUGCCGGCAA--- ------..(((((.(((.((...(((((........)))....(((((...((((((...-.)))))).)))))....---...))((.......)))).))).).))))..--- ( -26.60, z-score = 0.57, R) >apiMel3.Group10 4074145 108 + 11440700 CUUUACCACCUGUCUCGACUAUCACCGGCUCCCUCAG--CCACCAUCAUCAUCACCACCACGUGAGAACGCAUCACACCGCCUGCAACGUUCAACAGAGCAACGAACAGA----- .........((((.(((....((...((((.....))--))..........((((......))))(((((...((.......))...)))))....))....))))))).----- ( -19.40, z-score = -1.68, R) >consensus ______CAGCCGCCUCGACUCCAGCUCGAUAUUCGAGGACCUCGAUCGCUACUGCUGGCA_ACAGCAGCCGGUCACAA___CCAGCGCCAGCAAUGGUGGAACCGCCAGC_____ ................((((....(((.......)))..............((((((.....))))))..))))......................................... (-10.02 = -9.80 + -0.22)

| Location | 19,237,647 – 19,237,742 |
|---|---|
| Length | 95 |
| Sequences | 13 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 83.63 |
| Shannon entropy | 0.37030 |
| G+C content | 0.54901 |
| Mean single sequence MFE | -32.39 |
| Consensus MFE | -18.93 |
| Energy contribution | -18.11 |
| Covariance contribution | -0.82 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.907430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
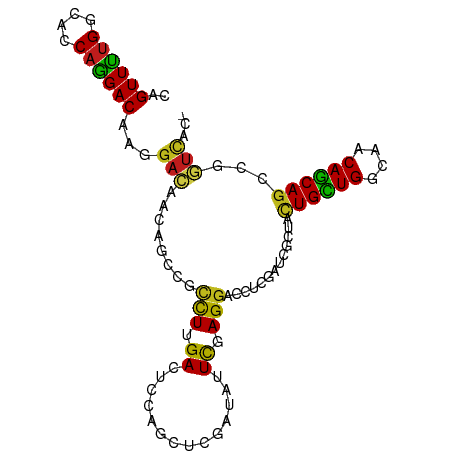
>dm3.chr3R 19237647 95 - 27905053 CAGUUUUGGCACCAGGACAAGGACAACAGUCGCCUUGACUCCAGCUCGAUUUUCGAGGACCUCGAUCGUUACUGCUGGCAACAGCAGCCGGUCAC- ..((((((....)))))).(((.....((((.....))))....((((.....))))..))).(((((...((((((....)))))).)))))..- ( -33.30, z-score = -2.41, R) >droSim1.chr3R 19055857 95 - 27517382 CAGUUUUGGCACCAGGACAAGGACAACAGUCGCCUCGACUCCAACUCGAUUUUCGAGGACCUCGAUCGCUACUGCUGGCAACAGCAGCCGGUCAC- ..((((((....)))))).(((.....(((((...)))))....((((.....))))..))).(((((...((((((....)))))).)))))..- ( -34.80, z-score = -3.23, R) >droSec1.super_0 19600617 95 - 21120651 CAGUUUUGGCACCAGGACAAGGACAACAGUCGCCUUGACUCCAACUCGAUUUUCGAGGACCUCGAUCGCUACUGCUGGCAACAGCAGUCGGUCAC- ..((((((....)))))).(((.....((((.....))))....((((.....))))..))).(((((..(((((((....))))))))))))..- ( -35.20, z-score = -3.30, R) >droYak2.chr3R 20117650 95 - 28832112 CAGUUUUGGCACCAGGACAAGGACAACAGCCGCCUGGACUCCAGCUCGAUAUUCGAGGACCUCGAUCGCUACUGCUGGCAACAGCAGCCGGUCAC- .....((((..(((((....((.(....))).)))))...))))((((.....))))......(((((...((((((....)))))).)))))..- ( -35.40, z-score = -2.65, R) >droEre2.scaffold_4820 1657853 95 + 10470090 CAGUUUUGGCAUCAGGACAAGGACAGCAGCCGCCUUGACUCCAACUCGAUAUUCGAGGACCUCGAUCGCUACUGCUGGCAACAGCAGUCGGUCAC- .......(((....((((((((.(.......))))))..)))..((((.....))))).))..(((((..(((((((....))))))))))))..- ( -34.00, z-score = -2.64, R) >droAna3.scaffold_13340 3380576 96 + 23697760 CAGUUCUGGCACCAGGACAAGGACAACAGCCGCCUCGACUCCAGCUCUUUCUUCGAGGAUCUGGAUCGCUACUGCUGGCAACAGCAAACUGUAACU ..((((((....)))))).......((((......(((.(((((.(((((....))))).))))))))....(((((....)))))..)))).... ( -33.30, z-score = -2.61, R) >dp4.chr2 2719292 95 - 30794189 CAGUUCUGGAACCAGGACAAGGACAGCAGUCGCCUCGAUUCUACCUCCAUAUUCGAGGACCUCGAUCGUUACUGCUGGCAACAGCAGCCGGUCAC- ..((((((....))))))...(((....))).((((((..............)))))).....(((((...((((((....)))))).)))))..- ( -36.84, z-score = -3.72, R) >droPer1.super_7 2812480 95 + 4445127 CAGUUCUGGAACCAGGACAAGGACAGCAGUCGCCUCGAUUCUACCUCCAUAUUCGAGGACCUCGAUCGUUACUGCUGGCAACAGCAGCCGGUCAC- ..((((((....))))))...(((....))).((((((..............)))))).....(((((...((((((....)))))).)))))..- ( -36.84, z-score = -3.72, R) >droWil1.scaffold_181130 9272984 95 - 16660200 CAGUUUUGGCAUCAAGACAAGGAUAACAGCCGUUUAGAUUCUAGUUCCAUUUUCGAGGAUCUGGAUCGCUAUUGUUGGCAGCAACAGCCAAUUGG- ..((((((....))))))...(((((.(((.(((((((((((.(.........).))))))))))).))).)))))(((.......)))......- ( -25.20, z-score = -1.62, R) >droVir3.scaffold_13047 8871271 95 - 19223366 CAGUUCUGGCAUCAGGACAAGGAUAACAGCCGCCUUGACUCCAGCUCUAUCUUCGAGGACCUUGAUCGAUACUGCUGGCAGCAGCAGCCGAUCAC- ..((((((....))))))...(((....((.((..((.(((((((..((((.(((((...)))))..))))..))))).)))))).))..)))..- ( -28.60, z-score = -0.72, R) >droMoj3.scaffold_6540 2348700 95 + 34148556 UAGUUUUGGCAUCAGGACAAGGACAGCAGCCGCCUGGACUCGAGCUCGAUUUUCGAGGACCUCGAUCGCUACUGCUGGCAGCAGCAGCCGAUCAC- .....(((((..((((....((.(....))).))))((.(((((((((.....))))...)))))))(((.(((....))).))).)))))....- ( -31.50, z-score = -0.70, R) >droGri2.scaffold_15074 6980856 95 + 7742996 CAGUUCUGGCACCAGGACAAGGACAGCAGCCGCCUUGACUCCAGCUCCAUAUUUGAGGACCUCGAUCGAUACUGUUGGCAGCAGCAGCCGAUCAC- ..((((((....))))))..(((...(((.....)))..)))((.(((........))).)).(((((...((((((....)))))).)))))..- ( -26.70, z-score = -0.20, R) >anoGam1.chr2R 60259253 92 + 62725911 CAGUUCUGGGGUCCGGACAAAGAUGCCAACCGGUUGGAUCCGG---CCAUGUUUGAGGAUCUGAAUCGCUAUUGUUGGAUACAGCAGAGCAACAC- ..((((..((((((...((((.(((....((((......))))---.))).)))).)))))))))).(((.((((((....))))))))).....- ( -29.40, z-score = -1.11, R) >consensus CAGUUUUGGCACCAGGACAAGGACAACAGCCGCCUUGACUCCAGCUCGAUAUUCGAGGACCUCGAUCGCUACUGCUGGCAACAGCAGCCGGUCAC_ ..((((((....))))))...(((........(((.((..............)).))).............((((((....))))))...)))... (-18.93 = -18.11 + -0.82)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:37:50 2011