| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,225,684 – 19,225,784 |
| Length | 100 |
| Max. P | 0.996026 |

| Location | 19,225,684 – 19,225,784 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 86.29 |
| Shannon entropy | 0.23918 |
| G+C content | 0.52318 |
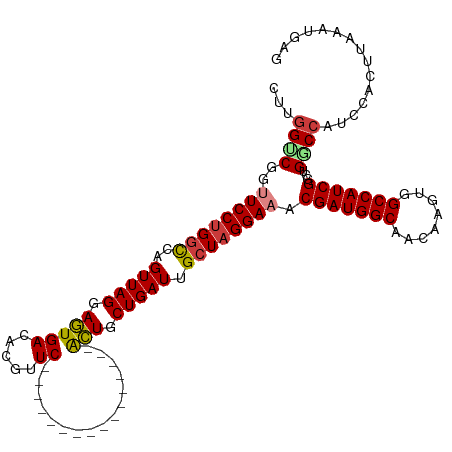
| Mean single sequence MFE | -42.54 |
| Consensus MFE | -29.56 |
| Energy contribution | -29.44 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.56 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.996026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
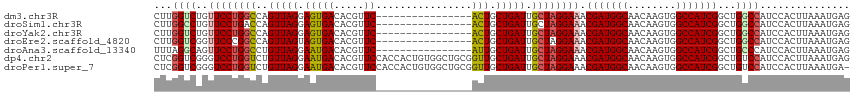
>dm3.chr3R 19225684 100 + 27905053 CUUGGUCUGUUCCUGGCCAGUUAGGAGUGACACGUUC----------------ACUGCUGAUUGCUAGGAAACGAUGGCAACAAGUGGCCAUCGGCUGGCCAUCCACUUAAAUGAG ..(((((.(((((((((.((((((.(((((.....))----------------))).)))))))))))))..(((((((........))))))))).))))).............. ( -43.40, z-score = -4.28, R) >droSim1.chr3R 19044027 100 + 27517382 CUUGGCCUGUUCCUGACCAGUUAGGAGUGACACGUUC----------------ACUGCUGAUUGCUAGGAAACGAUGGCAACAAGUGGCCAUCGGCUGGCCAUCCACUUAAAUGAG ..(((((.(((((((..(((((((.(((((.....))----------------))).))))))).)))))..(((((((........))))))))).))))).............. ( -41.40, z-score = -4.08, R) >droYak2.chr3R 20105566 100 + 28832112 CUUGGUCUGUUCCUGGCCAGUUAGGAGUGACACGUUC----------------ACUGCUGAUUGCUAGGAAACGAUGGCAACAAGUGGCCAUCGGCUGGCCAUCCACUUAAAUGAG ..(((((.(((((((((.((((((.(((((.....))----------------))).)))))))))))))..(((((((........))))))))).))))).............. ( -43.40, z-score = -4.28, R) >droEre2.scaffold_4820 1646121 100 - 10470090 CUUGGUCGGUUCCCGGCCAGUUAGUAGUGACACGUUC----------------ACUGCUGAUUGCUAGGAAACGAUGGCAACAAGUGGCCAUCGGCUGGCCAUCCACUUAAAUGAG ..(((((((((((..((.((((((((((((.....))----------------))))))))))))..)))..(((((((........))))))))))))))).............. ( -46.10, z-score = -5.09, R) >droAna3.scaffold_13340 3368291 100 - 23697760 UUUAGGCAGUUCCUGGCCUGUUAGGAAUGACACGUUC----------------AUUGCUGAUUGCUAGGAAACGAUGGCAACAAGUGGCCAUCGGCUGCCCAUCCACUUAAAUGAG ....(((((((((((((..(((((.(((((.....))----------------))).))))).)))))))..(((((((........)))))))))))))................ ( -42.70, z-score = -4.88, R) >dp4.chr2 2706750 116 + 30794189 CUCGGUCGGGUCCUGGUCUGUUAGGAAUGACACGUUCCACCACUGUGGCUGCGGUUGCUGAUUGCUAGGAAACGAUGGCAACAAGUGGCCAUCGGCUGUCCAUCCACUUAAAUGAG ...((.(((.(((((((..(((((.(((....(((.((((....))))..)))))).))))).)))))))..(((((((........))))))).))).))............... ( -40.40, z-score = -1.14, R) >droPer1.super_7 2799903 115 - 4445127 CUCGGUCGGGUCCUGGUCUGUUAGGAAUGACACGUUCCACCACUGUGGCUGCGGUUGCUGAUUGCUAGGAAACGAUGGCAACAAGUGGCCAUCGGCUGUCCAUCCACUUAAAUGA- ...((.(((.(((((((..(((((.(((....(((.((((....))))..)))))).))))).)))))))..(((((((........))))))).))).))..............- ( -40.40, z-score = -1.15, R) >consensus CUUGGUCGGUUCCUGGCCAGUUAGGAGUGACACGUUC________________ACUGCUGAUUGCUAGGAAACGAUGGCAACAAGUGGCCAUCGGCUGGCCAUCCACUUAAAUGAG ...((((..((((((((..(((((.(((.........................))).))))).)))))))).(((((((........)))))))...))))............... (-29.56 = -29.44 + -0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:37:46 2011