| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,212,023 – 19,212,131 |
| Length | 108 |
| Max. P | 0.500000 |

| Location | 19,212,023 – 19,212,131 |
|---|---|
| Length | 108 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 56.05 |
| Shannon entropy | 0.82383 |
| G+C content | 0.45889 |
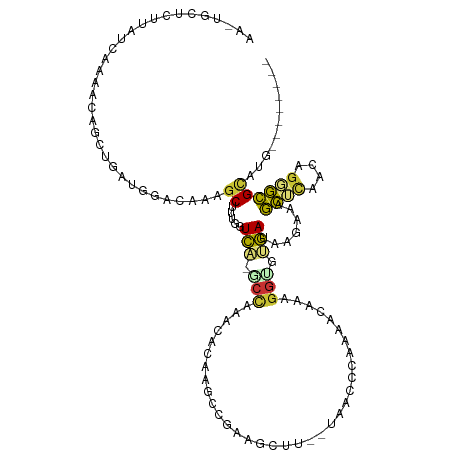
| Mean single sequence MFE | -28.21 |
| Consensus MFE | -3.92 |
| Energy contribution | -3.79 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.83 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.14 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
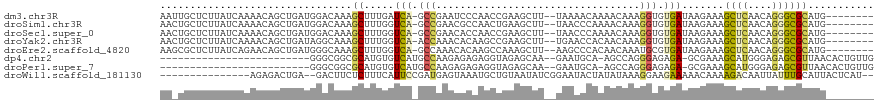
>dm3.chr3R 19212023 108 - 27905053 AAUUGCUCUUAUCAAAACAGCUGAUGGACAAAGCUUUGAUCA-GCCGAAUCCCAACCGAAGCUU--UAAAACAAAACAAAGGUGUGAUAAGAAAGCUCAACAGGGCGCAUG-------- ...(((((((((((...(.((((((.((.......)).))))-)).).......(((.......--..............))).))))))))..((((....)))))))..-------- ( -25.30, z-score = -1.48, R) >droSim1.chr3R 19030691 108 - 27517382 AACUGCUCUUAUCAAAACAGCUGAUGGACAAAGCUUUGGUCA-GCCGAACGCCAACUGAAGCUU--UAACCCAAAACAAAGGUGUGAUAAGAAAGCUCAACAGGGCGCAUG-------- ...((((((((((...(((.((..(((..(((((((..((..-((.....))..))..))))))--)...)))......)).))))))))))..((((....)))))))..-------- ( -32.70, z-score = -2.70, R) >droSec1.super_0 19575706 108 - 21120651 AACUGCUCUUAUCAAAACAGCUGAUGGACAAAGCUUUGGUCA-GCCGAACACCAACCGAAGCUU--UAACCCAAAACAAAGGUGUGAUAAGAAAGCUCAACAGGGCGCAUG-------- ...((((((((((...(((.((..(((..(((((((((((..-(.......)..))))))))))--)...)))......)).))))))))))..((((....)))))))..-------- ( -31.70, z-score = -2.86, R) >droYak2.chr3R 20091506 108 - 28832112 AACUGCUCUUAUCAAAACAGCUGAUAGGCAAAGCUUUGGUCA-ACCAAACACAAGCCGAAGCUU--UGAACCACAACAAAGGUGUGAUAAGAAAGCUCAACAGGGCGCAUG-------- ...(((((((((((..............((((((((((((..-...........))))))))))--)).(((........))).))))))))..((((....)))))))..-------- ( -33.52, z-score = -3.31, R) >droEre2.scaffold_4820 1632260 108 + 10470090 AAGCGCUCUUAUCAGAACAGCUGAUGGGCAAAGCUUUGGUCA-GCCAAACACAAGCCAAAGCUU--AAGCCCACAACAAAUGCGUGAUAAGAAAGCUCAACAGGGCGCAUG-------- ..((..((((((((...((..((.(((((.((((((((((..-(.......)..))))))))))--..)))))...))..))..))))))))..((((....))))))...-------- ( -37.30, z-score = -3.30, R) >dp4.chr2 2693892 90 - 30794189 -------------------------GGGCGGCGCAUGUGUCAUGCCAAGAGAGAGGUAGAGCAA--GAAUGCA-AGCCAGGGAGAGA-GCGAAAGCAUGGGAGAGCGUUAACACUGUUG -------------------------..(((((((((.(((..((((........))))..))).--..)))).-.))).........-((....))........))............. ( -24.10, z-score = -1.23, R) >droPer1.super_7 2786898 90 + 4445127 -------------------------GGGCGGCGCAUGUGUCAUGCCAAGAGAGAGGUAGAGCAA--GAAUGCA-AGCCAGGGAGAGA-GCGAAAGCAUGGGAGAGCGUUAACACUGUUG -------------------------..(((((((((.(((..((((........))))..))).--..)))).-.))).........-((....))........))............. ( -24.10, z-score = -1.23, R) >droWil1.scaffold_181130 9243785 100 - 16660200 ---------------AGAGACUGA--GACUUCUCUUUCAUUCCGAUGAGUAAAUGCUGUAAUAUCGGAAUACUAUAUAAAGGAAGAAAAACAAAAGACAAUUAUUUGCAUUACUCAU-- ---------------......(((--(.((((.((((.((((((((((((....)))....))))))))).......))))))))..........(.(((....))))....)))).-- ( -17.00, z-score = -0.87, R) >consensus AA_UGCUCUUAUCAAAACAGCUGAUGGACAAAGCUUUGGUCA_GCCAAACACAAGCCGAAGCUU__UAACCCAAAACAAAGGUGUGAUAAGAAAGCUCAACAGGGCGCAUG________ ................................((.....(((.(((..................................))).))).......((((....))))))........... ( -3.92 = -3.79 + -0.13)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:37:41 2011