
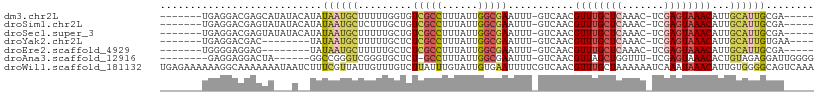
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,417,699 – 8,417,794 |
| Length | 95 |
| Max. P | 0.992110 |

| Location | 8,417,699 – 8,417,794 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 70.88 |
| Shannon entropy | 0.54571 |
| G+C content | 0.40266 |
| Mean single sequence MFE | -24.67 |
| Consensus MFE | -15.52 |
| Energy contribution | -16.43 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.983044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
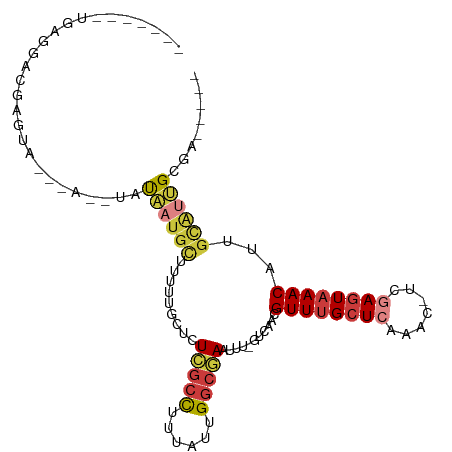
>dm3.chr2L 8417699 95 + 23011544 -------UGAGGACGAGCAUAUACAUAUAAUGCUUUUUGGUGUCGCCUUUAUUGGCGAAUUU-GUCAACGUUUGCUCAAAC-UCGAGUAAACAUUGCAUUGCGA----- -------....(((((((((.........)))))).......(((((......)))))....-)))...((((((((....-..)))))))).((((...))))----- ( -26.30, z-score = -1.84, R) >droSim1.chr2L 8203833 95 + 22036055 -------UGAGGACGAGUAUAUACAUAUAAUGCUCUUUGCUGUCGCCUUUAUUGGCGAAUUU-GUCAACGUUUGCUCAAAC-UCGAGUAAACAUUGCAUUGCGA----- -------.......((((((.........)))))).((((..(((((......)))))...(-(.(((.((((((((....-..)))))))).)))))..))))----- ( -27.70, z-score = -2.58, R) >droSec1.super_3 3914002 95 + 7220098 -------UGAGGACGAGUAUAUACAUAUAAUGCUUUUUGCUGUCGCCUUUAUUGGCGAAUUU-GUCAACGUUUGCUCAAAC-UCGAGUAAACAUUGCAUUGCGA----- -------....(((((((.............((.....))..(((((......)))))))))-)))...((((((((....-..)))))))).((((...))))----- ( -26.80, z-score = -2.31, R) >droYak2.chr2L 11055369 88 + 22324452 -------UGAGGACGAC--------UAUAAUGCUUUUUGCUCUCGCCUUUAUUGGCGAAUUU-GUCAACGUUUGCUCAAAC-UCGAGUAAACAUUGCAUUGUGAA---- -------..........--------((((((((.........(((((......)))))....-......((((((((....-..))))))))...))))))))..---- ( -26.50, z-score = -3.42, R) >droEre2.scaffold_4929 17326449 87 + 26641161 -------UGGGGAGGAG--------UAUAAUGCUUUUUGCUCUCGCCUUUAUUGGCGAAUUU-GUCAACGUUUGCUCAAAC-UCGAGUAAACAUUGCAUUGCGA----- -------.((.((((((--------(.....))))))).)).(((((......)))))...(-(.(((.((((((((....-..)))))))).)))))......----- ( -26.60, z-score = -2.38, R) >droAna3.scaffold_12916 10074709 92 + 16180835 --------GAGGAGGACUA------GGCCGGGUCGGGUGCUCU-GCCUUUAUUGGCGAAUUU-GUCAACGUUAGCUGGUUU-UCGAGUAAACACUGUAGAGGAUUGGGG --------..((((.(((.------(((...))).))).))))-.((((((((((((....)-))))..(((.(((.(...-.).))).)))...)))))))....... ( -23.70, z-score = 0.68, R) >droWil1.scaffold_181132 307044 109 + 1035393 UGAGAAAAAAGGCAAAAAAAUAAUCUUUCGUUAUUGUUUGUCUUAUUUGUAUUGUGAAUUUUCGUCAACGUUUGCUAAAAAAUCAAAUAAACAUUGUGGGGCAGUCAAA ..(.(((.((((((((..((((((.....)))))).)))))))).))).)....(((..(..((.(((.(((((.............))))).)))))..)...))).. ( -15.12, z-score = 0.30, R) >consensus _______UGAGGACGAGUA___A__UAUAAUGCUUUUUGCUCUCGCCUUUAUUGGCGAAUUU_GUCAACGUUUGCUCAAAC_UCGAGUAAACAUUGCAUUGCGA_____ ...........................((((((.........(((((......)))))...........((((((((.......))))))))...))))))........ (-15.52 = -16.43 + 0.90)
| Location | 8,417,699 – 8,417,794 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 70.88 |
| Shannon entropy | 0.54571 |
| G+C content | 0.40266 |
| Mean single sequence MFE | -20.03 |
| Consensus MFE | -14.00 |
| Energy contribution | -14.43 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.992110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
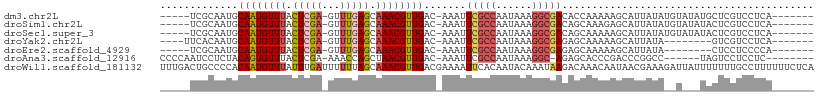
>dm3.chr2L 8417699 95 - 23011544 -----UCGCAAUGCAAUGUUUACUCGA-GUUUGAGCAAACGUUGAC-AAAUUCGCCAAUAAAGGCGACACCAAAAAGCAUUAUAUGUAUAUGCUCGUCCUCA------- -----..(((.((((((((((.((((.-...)))).)))))))).)-)...(((((......))))).......................))).........------- ( -22.40, z-score = -1.80, R) >droSim1.chr2L 8203833 95 - 22036055 -----UCGCAAUGCAAUGUUUACUCGA-GUUUGAGCAAACGUUGAC-AAAUUCGCCAAUAAAGGCGACAGCAAAGAGCAUUAUAUGUAUAUACUCGUCCUCA------- -----..((..((((((((((.((((.-...)))).)))))))).)-)...(((((......)))))..))...(((...(((....)))..))).......------- ( -23.50, z-score = -1.98, R) >droSec1.super_3 3914002 95 - 7220098 -----UCGCAAUGCAAUGUUUACUCGA-GUUUGAGCAAACGUUGAC-AAAUUCGCCAAUAAAGGCGACAGCAAAAAGCAUUAUAUGUAUAUACUCGUCCUCA------- -----..((..((((((((((.((((.-...)))).)))))))).)-)...(((((......)))))..))...............................------- ( -22.10, z-score = -1.79, R) >droYak2.chr2L 11055369 88 - 22324452 ----UUCACAAUGCAAUGUUUACUCGA-GUUUGAGCAAACGUUGAC-AAAUUCGCCAAUAAAGGCGAGAGCAAAAAGCAUUAUA--------GUCGUCCUCA------- ----.....((((((((((((.((((.-...)))).)))))))...-...((((((......))))))........)))))...--------..........------- ( -21.80, z-score = -2.33, R) >droEre2.scaffold_4929 17326449 87 - 26641161 -----UCGCAAUGCAAUGUUUACUCGA-GUUUGAGCAAACGUUGAC-AAAUUCGCCAAUAAAGGCGAGAGCAAAAAGCAUUAUA--------CUCCUCCCCA------- -----..((..((((((((((.((((.-...)))).)))))))).)-)..((((((......)))))).)).............--------..........------- ( -23.10, z-score = -3.17, R) >droAna3.scaffold_12916 10074709 92 - 16180835 CCCCAAUCCUCUACAGUGUUUACUCGA-AAACCAGCUAACGUUGAC-AAAUUCGCCAAUAAAGGC-AGAGCACCCGACCCGGCC------UAGUCCUCCUC-------- ...............((((((......-....((((....))))..-......(((......)))-.))))))..(((......------..)))......-------- ( -15.90, z-score = -1.00, R) >droWil1.scaffold_181132 307044 109 - 1035393 UUUGACUGCCCCACAAUGUUUAUUUGAUUUUUUAGCAAACGUUGACGAAAAUUCACAAUACAAAUAAGACAAACAAUAACGAAAGAUUAUUUUUUUGCCUUUUUUCUCA .............((((((((..((((....)))).))))))))..(((((..............(((.((((.((((((....).)))))..)))).))))))))... ( -11.43, z-score = -0.02, R) >consensus _____UCGCAAUGCAAUGUUUACUCGA_GUUUGAGCAAACGUUGAC_AAAUUCGCCAAUAAAGGCGAGAGCAAAAAGCAUUAUA__U___UACUCGUCCUCA_______ .............((((((((.((((.....)))).)))))))).......(((((......))))).......................................... (-14.00 = -14.43 + 0.43)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:25:54 2011