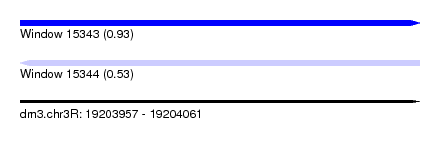
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,203,957 – 19,204,061 |
| Length | 104 |
| Max. P | 0.934159 |

| Location | 19,203,957 – 19,204,061 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 69.17 |
| Shannon entropy | 0.62032 |
| G+C content | 0.49268 |
| Mean single sequence MFE | -29.24 |
| Consensus MFE | -14.25 |
| Energy contribution | -13.82 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.934159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
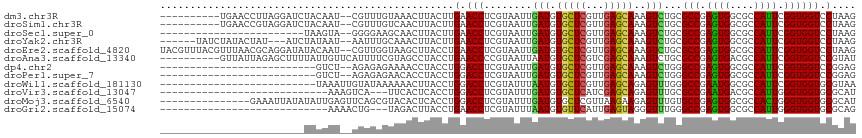
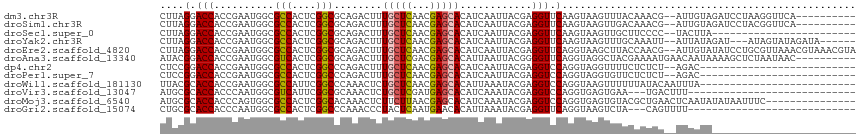
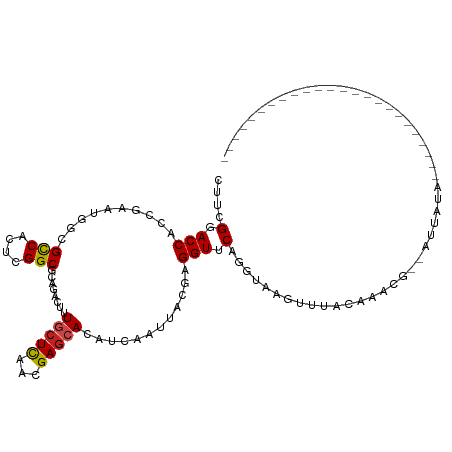
>dm3.chr3R 19203957 104 + 27905053 ----------UGAACCUUAGGAUCUACAAU--CGUUUGUAAACUUACUUGAACCUCGUAAUUGAUGUGCUCGUUGAGCAAAGUCUGCGCCGAGUGGCGCCAUUCGGUGGUCCUAAG ----------.....(((((((((......--.....(((.((((......((.(((....))).))((((...)))).)))).)))((((((((....))))))))))))))))) ( -33.80, z-score = -3.07, R) >droSim1.chr3R 19022823 104 + 27517382 ----------UGAACCGUAGGAUCUACAAU--CGUUUGUCAACUUACUUGAACCUCGUAAUUGAUGUGCUCGUUGAGCAAAGUCUGCGCCGAGUGGCGCCAUUCGGUGGUCCUAAG ----------.......(((((((..((((--.((.((((((.((((.........)))))))))).))..)))).(((.....)))((((((((....))))))))))))))).. ( -30.80, z-score = -1.58, R) >droSec1.super_0 19567897 90 + 21120651 ------------------------UAAGUA--GGGGAAGCAACUUACUUGAACCUCGUAAUUGAUGUGCUCGUUGAGCAAAGUCUGCGCCGAGUGGCGCCAUUCGGUGGUCCUAAG ------------------------((((((--((........)))))))).(((..(((...(((.(((((...)))))..))))))((((((((....)))))))))))...... ( -28.50, z-score = -1.25, R) >droYak2.chr3R 20083334 105 + 28832112 ------UAUCUAUACUAU---AUCUAUAAU--AAUUUGCAAACUUACUUGAACCUCGUAAUUGAUGUGCUCGUUGAGCAAAGUCUGCGCCGAGUGGCGCCAUUCGGUGGUCCUAAG ------............---.........--.....(((.((((......((.(((....))).))((((...)))).)))).)))((((((((....))))))))......... ( -23.90, z-score = -1.00, R) >droEre2.scaffold_4820 1624152 114 - 10470090 UACGUUUACGUUUAACGCAGGAUAUACAAU--CGUUGGUAAGCUUACCUGAACCUCGUAAUUGAUGUGCUCGUUGAGCAAAGUCUGCGCCGAGUGGCGCCAUUCGGUGGUCCUAAG .........((((((((.((.((((.((((--((..(((.((.....))..))).))..)))))))).))))))))))........(((((((((....)))))))))........ ( -33.80, z-score = -1.06, R) >droAna3.scaffold_13340 3347014 106 - 23697760 ----------GUUAUUAGAGCUUUUAUUGUUCAUUUUCGUAGCCUACCUGAACCCCGUAAUUAAUGUGCUCGUCGAGCAAAGUCUGCGCCGAGUGACGCCAUUCGGUGGUCCGUAU ----------.......((((.......)))).........((..(((........(((.......(((((...))))).....)))((((((((....)))))))))))..)).. ( -23.60, z-score = -0.28, R) >dp4.chr2 2685035 88 + 30794189 --------------------------GUCU--AGAGAGAAAACCUACCUGGACCUCGUAAUUGAUGUGCUCGUUGAGCAAAGUCUGGGCCGAGUGGCGCCAUUCGGUGGUCCGGAG --------------------------....--...............(((((((........(((.(((((...)))))..)))...((((((((....))))))))))))))).. ( -31.40, z-score = -1.73, R) >droPer1.super_7 2777864 88 - 4445127 --------------------------GUCU--AGAGAGAACACCUACCUGGACCUCGUAAUUGAUGUGCUCGUUGAGCAAAGUCUGGGCCGAGUGGCGCCAUUCGGUGGUCCGGAG --------------------------....--...............(((((((........(((.(((((...)))))..)))...((((((((....))))))))))))))).. ( -31.40, z-score = -1.32, R) >droWil1.scaffold_181130 9235616 90 + 16660200 --------------------------UAAAUUGUAUAAAAAACUUACCUGGACCUCGUAUUUAAUGUGCUCGUUGAGCAGAGUUUGGGCCGAAUGGCGCCAUUCGGUGGUGCGUAA --------------------------......((((..........((..(((.((...(((((((....)))))))..)))))..))(((((((....)))))))..)))).... ( -25.40, z-score = -1.25, R) >droVir3.scaffold_13047 8838279 85 + 19223366 ----------------------------AAAGUCA---UUCACUCACCUGGACCUCGUAUUUGAUGUGCUCAUCGAGCAGAGUUUGCGCCGAAUGACGCCAUUGGGUGGUGCGCAU ----------------------------.......---..(((.((((..(..(((...(((((((....)))))))..)))...(((........)))..)..)))))))..... ( -21.80, z-score = 0.80, R) >droMoj3.scaffold_6540 2306664 101 - 34148556 ---------------GAAAUUAUAUAUUGAGUUCAGCGUACACUCACCUGGACCUCGUAUUUGAUGUGCUCGUUAAGAAGAGUUUGUGCCGAGUGGCGCCACUGGGUGGUGCGCAU ---------------(((.(((.....))).))).((((((...((((..(..(((...(((((((....)))))))..)))...(((((....)))))..)..)))))))))).. ( -33.00, z-score = -1.66, R) >droGri2.scaffold_15074 6950172 85 - 7742996 ----------------------------AAAACUG---UAGACUUACCUGAACCUCGUAUUUAAUGUGUUCAUUGAGUAGGGUUUGGGCCGAGUGGCGCCAUUGGGUGGUGCGCAG ----------------------------....(((---(..((((.((..(((((..(((((((((....))))))))))))))..))..)))).(((((((...))))))))))) ( -33.50, z-score = -3.51, R) >consensus ________________________UAUAAU__AGAUAGUAAACUUACCUGAACCUCGUAAUUGAUGUGCUCGUUGAGCAAAGUCUGCGCCGAGUGGCGCCAUUCGGUGGUCCGAAG .................................................(.(((........(((.(((((...)))))..)))...(((.((((....)))).)))))).).... (-14.25 = -13.82 + -0.43)
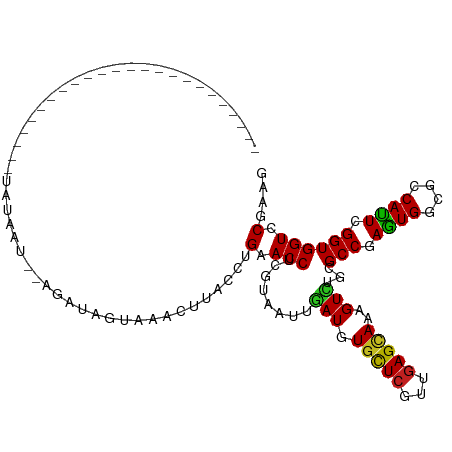
| Location | 19,203,957 – 19,204,061 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 69.17 |
| Shannon entropy | 0.62032 |
| G+C content | 0.49268 |
| Mean single sequence MFE | -24.12 |
| Consensus MFE | -10.70 |
| Energy contribution | -10.73 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.533167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19203957 104 - 27905053 CUUAGGACCACCGAAUGGCGCCACUCGGCGCAGACUUUGCUCAACGAGCACAUCAAUUACGAGGUUCAAGUAAGUUUACAAACG--AUUGUAGAUCCUAAGGUUCA---------- (((((((..(((.....(((((....))))).((...(((((...)))))..))........))).........(((((((...--.)))))))))))))).....---------- ( -30.20, z-score = -2.60, R) >droSim1.chr3R 19022823 104 - 27517382 CUUAGGACCACCGAAUGGCGCCACUCGGCGCAGACUUUGCUCAACGAGCACAUCAAUUACGAGGUUCAAGUAAGUUGACAAACG--AUUGUAGAUCCUACGGUUCA---------- ....(((((..((....(((((....)))))....((((.(((((((((...((......)).))))......)))))))))))--...((((...))))))))).---------- ( -28.50, z-score = -1.49, R) >droSec1.super_0 19567897 90 - 21120651 CUUAGGACCACCGAAUGGCGCCACUCGGCGCAGACUUUGCUCAACGAGCACAUCAAUUACGAGGUUCAAGUAAGUUGCUUCCCC--UACUUA------------------------ ....(((((........(((((....))))).((...(((((...)))))..))........)))))(((((.(........).--))))).------------------------ ( -23.80, z-score = -1.39, R) >droYak2.chr3R 20083334 105 - 28832112 CUUAGGACCACCGAAUGGCGCCACUCGGCGCAGACUUUGCUCAACGAGCACAUCAAUUACGAGGUUCAAGUAAGUUUGCAAAUU--AUUAUAGAU---AUAGUAUAGAUA------ ....(((((........(((((....))))).((...(((((...)))))..))........)))))..(((....)))....(--(((((....---))))))......------ ( -23.50, z-score = -1.23, R) >droEre2.scaffold_4820 1624152 114 + 10470090 CUUAGGACCACCGAAUGGCGCCACUCGGCGCAGACUUUGCUCAACGAGCACAUCAAUUACGAGGUUCAGGUAAGCUUACCAACG--AUUGUAUAUCCUGCGUUAAACGUAAACGUA ....(((((........(((((....))))).((...(((((...)))))..))........))))).((((....))))....--...........((((((.......)))))) ( -29.10, z-score = -0.85, R) >droAna3.scaffold_13340 3347014 106 + 23697760 AUACGGACCACCGAAUGGCGUCACUCGGCGCAGACUUUGCUCGACGAGCACAUUAAUUACGGGGUUCAGGUAGGCUACGAAAAUGAACAAUAAAAGCUCUAAUAAC---------- ....(((((.(((....(((((....)))))......(((((...))))).........))))))))...((((((..................))).))).....---------- ( -26.67, z-score = -1.65, R) >dp4.chr2 2685035 88 - 30794189 CUCCGGACCACCGAAUGGCGCCACUCGGCCCAGACUUUGCUCAACGAGCACAUCAAUUACGAGGUCCAGGUAGGUUUUCUCUCU--AGAC-------------------------- (((((((((......(((.(((....))))))((...(((((...)))))..))........))))).)).))...........--....-------------------------- ( -25.40, z-score = -1.27, R) >droPer1.super_7 2777864 88 + 4445127 CUCCGGACCACCGAAUGGCGCCACUCGGCCCAGACUUUGCUCAACGAGCACAUCAAUUACGAGGUCCAGGUAGGUGUUCUCUCU--AGAC-------------------------- (((((((((......(((.(((....))))))((...(((((...)))))..))........))))).)).))...........--....-------------------------- ( -25.40, z-score = -0.68, R) >droWil1.scaffold_181130 9235616 90 - 16660200 UUACGCACCACCGAAUGGCGCCAUUCGGCCCAAACUCUGCUCAACGAGCACAUUAAAUACGAGGUCCAGGUAAGUUUUUUAUACAAUUUA-------------------------- ..((..(((.(((((((....)))))))......((((((((...)))))..........))).....)))..))...............-------------------------- ( -19.40, z-score = -1.46, R) >droVir3.scaffold_13047 8838279 85 - 19223366 AUGCGCACCACCCAAUGGCGUCAUUCGGCGCAAACUCUGCUCGAUGAGCACAUCAAAUACGAGGUCCAGGUGAGUGAA---UGACUUU---------------------------- .....(((((((.....(((((....)))))...((((((((...)))))..........))).....)))).)))..---.......---------------------------- ( -21.70, z-score = 0.21, R) >droMoj3.scaffold_6540 2306664 101 + 34148556 AUGCGCACCACCCAGUGGCGCCACUCGGCACAAACUCUUCUUAACGAGCACAUCAAAUACGAGGUCCAGGUGAGUGUACGCUGAACUCAAUAUAUAAUUUC--------------- ..((((((((((...(((.(((.((((.................))))....((......)))))))))))).)))..)))....................--------------- ( -20.33, z-score = 0.23, R) >droGri2.scaffold_15074 6950172 85 + 7742996 CUGCGCACCACCCAAUGGCGCCACUCGGCCCAAACCCUACUCAAUGAACACAUUAAAUACGAGGUUCAGGUAAGUCUA---CAGUUUU---------------------------- ..((..(((......(((.(((....))))))............(((((...((......)).))))))))..))...---.......---------------------------- ( -15.40, z-score = 0.03, R) >consensus CUUCGGACCACCGAAUGGCGCCACUCGGCGCAGACUUUGCUCAACGAGCACAUCAAUUACGAGGUUCAGGUAAGUUUACAAACG__AUUAUA________________________ ....(.(((..........(((....)))........(((((...)))))............))).)................................................. (-10.70 = -10.73 + 0.04)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:37:40 2011