
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,148,103 – 19,148,205 |
| Length | 102 |
| Max. P | 0.759763 |

| Location | 19,148,103 – 19,148,205 |
|---|---|
| Length | 102 |
| Sequences | 10 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 65.35 |
| Shannon entropy | 0.67527 |
| G+C content | 0.49330 |
| Mean single sequence MFE | -25.17 |
| Consensus MFE | -8.88 |
| Energy contribution | -7.63 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
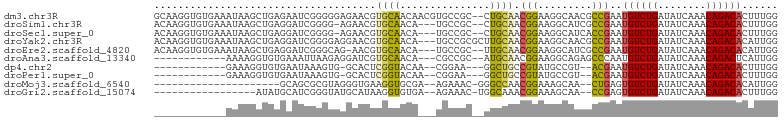
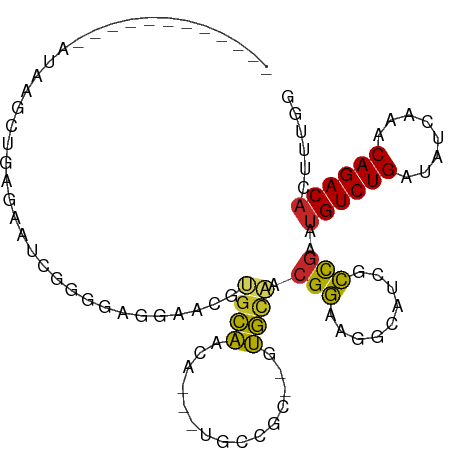
>dm3.chr3R 19148103 102 + 27905053 GCAAGGUGUGAAAUAAGCUGAGAAUCGGGGGAGAACGUGCAACAACGUGCCGC--CUGCAACGGAAGGCAACGCCGAAUGUCUGAUAUCAAACAGACACUUUGG .((((((((...............((((.((...((((......)))).))((--((........))))....)))).(((.((....)).))).)))))))). ( -30.30, z-score = -1.35, R) >droSim1.chr3R 18964952 98 + 27517382 ACAAGGUGUGAAAUAAGCUGAGGAUCGGGG-AGAACGUGCAACA---UGCCGC--CUGCAACGGAAGGCAUCGCCGAAUGUCUGAUAUCAAACAGACACUUUGG .((((((((...............((((.(-(....(.((....---.)))((--((........)))).)).)))).(((.((....)).))).)))))))). ( -28.30, z-score = -1.12, R) >droSec1.super_0 19512773 98 + 21120651 ACAAGGUGUGAAAUAAGCUGAGGAUCGGGG-AGAACGUGCAACA---UGCCGC--CUGCAACGGAAGGCAUCACCGAAUGUCUGAUAUCAAACAGACACUUUGG .((((((((...............((((.(-(....(.((....---.)))((--((........)))).)).)))).(((.((....)).))).)))))))). ( -26.20, z-score = -0.76, R) >droYak2.chr3R 20025275 101 + 28832112 ACAAGGUGUGAAAUAAGCUGAGGAUCGGGGAGGAACGUGCAACA---UGCCGCGCUUGCAACGGAAGGCAACGCCGAAUGUCUGAUAUCAAACAGACACAUUGG .((((((((........(((.....)))(.(((..((.((....---.))))..))).)..(....)...)))))...((((((........))))))..))). ( -24.20, z-score = 0.29, R) >droEre2.scaffold_4820 1564729 98 - 10470090 ACAAGGUGUGAAAUAAGCUGAGGAUCGGGCAG-AACGUGCAACA---UGCCGC--UUGCAACGGAAGGCAUCGCCGAAUGUCUGAUAUCAAACAGACACAUUGG ....((((((........(((..((((((((.-..((.((...(---((((..--...........))))).))))..)))))))).)))......)))))).. ( -25.26, z-score = -0.22, R) >droAna3.scaffold_13340 15459550 87 - 23697760 ------------AAAAGGUGUGAAAUUAAGAGGAUCGUGCAACA---CGCCGC--AUGCAACGGAAGGCAGAGCCCAAUGUCUGAUAUCAAACAGACUCAUUGG ------------....((((((.......(.....)......))---))))((--.(((..(....))))..))((((((((((........)))))..))))) ( -21.42, z-score = -0.83, R) >dp4.chr2 21311683 84 - 30794189 ------------GAAAGGUGUGAAUAAAGUG-GCACUCGGUACAA--CGGAA---GGCUGCCGUAUGCCGU--ACGAAUGUCUGAUAUCAAACAGACACUUUGG ------------............(((((((-(((..(((((...--(....---)..)))))..))))..--......(((((........))))))))))). ( -26.10, z-score = -2.43, R) >droPer1.super_0 10997019 84 + 11822988 ------------GAAAGGUGUGAAUAAAGUG-GCACUCGGUACAA--CGGAA---GGCUGCCGUAUGCCGU--ACGAAUGUCUGAUAUCAAACAGACACUUUGG ------------............(((((((-(((..(((((...--(....---)..)))))..))))..--......(((((........))))))))))). ( -26.10, z-score = -2.43, R) >droMoj3.scaffold_6540 10699502 78 + 34148556 ---------------------GCAGCGCGUAGGGUGAAGGUGCGA--AGAAAC-GGGCCAACGGAAAGCAA--CUGAGUGUCUGAUAUCAAACAGACACAUUGG ---------------------((..(((.....)))..(((.((.--.....)-).)))........))..--(..((((((((........))))))).)..) ( -21.30, z-score = -1.37, R) >droGri2.scaffold_15074 7135100 82 - 7742996 -----------------AUAUGCAUCGGGUAUGCAUAAGGUGUGA--AGAAAC-UGGCAAACGGAAAGCAA--CCGAGUGUCUGAUAUCAAACAGACACUUUGG -----------------.(((((((.....))))))).(((.((.--.....(-((.....)))....)))--))(((((((((........)))))))))... ( -22.50, z-score = -2.34, R) >consensus ____________AUAAGCUGAGAAUCGGGGAGGAACGUGCAACA___UGCCGC__GUGCAACGGAAGGCAUCGCCGAAUGUCUGAUAUCAAACAGACACUUUGG .....................................((((...............)))).(((.........)))..((((((........))))))...... ( -8.88 = -7.63 + -1.25)
| Location | 19,148,103 – 19,148,205 |
|---|---|
| Length | 102 |
| Sequences | 10 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 65.35 |
| Shannon entropy | 0.67527 |
| G+C content | 0.49330 |
| Mean single sequence MFE | -22.02 |
| Consensus MFE | -8.76 |
| Energy contribution | -8.13 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.759763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
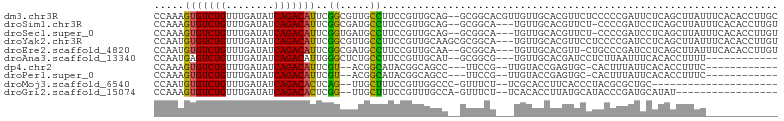
>dm3.chr3R 19148103 102 - 27905053 CCAAAGUGUCUGUUUGAUAUCAGACAUUCGGCGUUGCCUUCCGUUGCAG--GCGGCACGUUGUUGCACGUUCUCCCCCGAUUCUCAGCUUAUUUCACACCUUGC .(((.((((.((((((....)))))).((((.(((((((........))--)))))((((......))))......))))...............)))).))). ( -25.00, z-score = -0.69, R) >droSim1.chr3R 18964952 98 - 27517382 CCAAAGUGUCUGUUUGAUAUCAGACAUUCGGCGAUGCCUUCCGUUGCAG--GCGGCA---UGUUGCACGUUCU-CCCCGAUCCUCAGCUUAUUUCACACCUUGU .(((.((((.((((((....)))))).((((.(((((((........))--)))((.---....))......)-).))))...............)))).))). ( -22.10, z-score = 0.04, R) >droSec1.super_0 19512773 98 - 21120651 CCAAAGUGUCUGUUUGAUAUCAGACAUUCGGUGAUGCCUUCCGUUGCAG--GCGGCA---UGUUGCACGUUCU-CCCCGAUCCUCAGCUUAUUUCACACCUUGU ....((((((((........)))))))).(((((((((..((......)--).))))---)((((.....((.-....))....))))........)))).... ( -24.90, z-score = -0.97, R) >droYak2.chr3R 20025275 101 - 28832112 CCAAUGUGUCUGUUUGAUAUCAGACAUUCGGCGUUGCCUUCCGUUGCAAGCGCGGCA---UGUUGCACGUUCCUCCCCGAUCCUCAGCUUAUUUCACACCUUGU .(((.((((.((((((....)))))).((((((((((........)).)))))((.(---((.....))).))....)))...............)))).))). ( -22.00, z-score = -0.17, R) >droEre2.scaffold_4820 1564729 98 + 10470090 CCAAUGUGUCUGUUUGAUAUCAGACAUUCGGCGAUGCCUUCCGUUGCAA--GCGGCA---UGUUGCACGUU-CUGCCCGAUCCUCAGCUUAUUUCACACCUUGU .(((.((((.((((((....)))))).(((((((((.....))))))..--((((.(---((.....))).-)))).)))...............)))).))). ( -23.30, z-score = -0.24, R) >droAna3.scaffold_13340 15459550 87 + 23697760 CCAAUGAGUCUGUUUGAUAUCAGACAUUGGGCUCUGCCUUCCGUUGCAU--GCGGCG---UGUUGCACGAUCCUCUUAAUUUCACACCUUUU------------ ....((((((((........)))))...(((.(((((..(.(((((...--.)))))---.)..))).)).))).......)))........------------ ( -19.10, z-score = -0.26, R) >dp4.chr2 21311683 84 + 30794189 CCAAAGUGUCUGUUUGAUAUCAGACAUUCGU--ACGGCAUACGGCAGCC---UUCCG--UUGUACCGAGUGC-CACUUUAUUCACACCUUUC------------ ..((((((((((........)))))......--..(((((.(((((((.---....)--)))..))).))))-)))))).............------------ ( -23.40, z-score = -3.01, R) >droPer1.super_0 10997019 84 - 11822988 CCAAAGUGUCUGUUUGAUAUCAGACAUUCGU--ACGGCAUACGGCAGCC---UUCCG--UUGUACCGAGUGC-CACUUUAUUCACACCUUUC------------ ..((((((((((........)))))......--..(((((.(((((((.---....)--)))..))).))))-)))))).............------------ ( -23.40, z-score = -3.01, R) >droMoj3.scaffold_6540 10699502 78 - 34148556 CCAAUGUGUCUGUUUGAUAUCAGACACUCAG--UUGCUUUCCGUUGGCCC-GUUUCU--UCGCACCUUCACCCUACGCGCUGC--------------------- .....(((((((........))))))).(((--(.((.....((.((..(-(.....--.))..))...)).....)))))).--------------------- ( -16.20, z-score = -1.08, R) >droGri2.scaffold_15074 7135100 82 + 7742996 CCAAAGUGUCUGUUUGAUAUCAGACACUCGG--UUGCUUUCCGUUUGCCA-GUUUCU--UCACACCUUAUGCAUACCCGAUGCAUAU----------------- ....((((((((........)))))))).((--(.((.....))..))).-......--........(((((((.....))))))).----------------- ( -20.80, z-score = -3.10, R) >consensus CCAAAGUGUCUGUUUGAUAUCAGACAUUCGGCGAUGCCUUCCGUUGCAC__GCGGCA___UGUUGCACGUUCCUCCCCGAUCCUCAGCUUAU____________ .....(((((((........)))))))..((.....)).................................................................. ( -8.76 = -8.13 + -0.63)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:37:34 2011