| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,408,166 – 8,408,271 |
| Length | 105 |
| Max. P | 0.963893 |

| Location | 8,408,166 – 8,408,271 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 71.41 |
| Shannon entropy | 0.59610 |
| G+C content | 0.40633 |
| Mean single sequence MFE | -15.40 |
| Consensus MFE | -8.53 |
| Energy contribution | -8.73 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.963893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
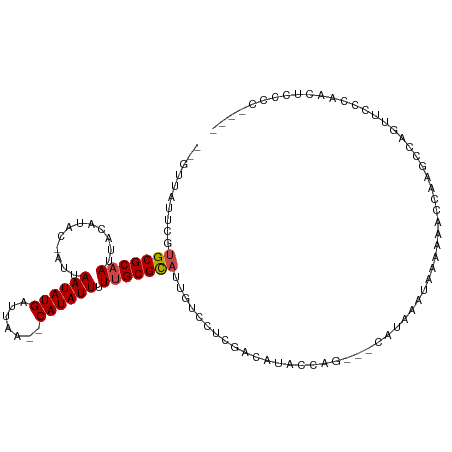
>dm3.chr2L 8408166 105 + 23011544 --GUUAUUCGUGCGCAAUUACAUAC-AUUAAUAUGAUUAA--CAUAUUUUUGCGCAUUGUCCUCGACAUACCAG---CAUAAAUAAGAUGCCCAGCCAGUUCCCAACUCCCGC--- --.......((((((((........-...((((((.....--)))))).))))))))((((...)))).....(---(((.......))))......................--- ( -15.50, z-score = -0.73, R) >droSim1.chr2L 8194319 105 + 22036055 --GUUAUUCGUGCGCAAUUACAUAC-AUUAAUAUGAUUAA--CAUAUUUUUGCGCAUUGUCCUCGACAUACCAG---CAUAAAUAAGAAGCCCAGCCAGUUCCCAACUUCCCG--- --....(((((((((((........-...((((((.....--)))))).))))))))((((...))))......---.........)))........................--- ( -14.40, z-score = -1.10, R) >droSec1.super_3 3904560 105 + 7220098 --GUUAUUCGUGCGCAAUUACAUAC-AUUAAUAUGAUUAA--CAUAUUUUUGCGCAUUGUCCUCGACAUACCAG---CAUAAAUAAGAAGCCCAGCCAGUUCCCAACUCCCCG--- --....(((((((((((........-...((((((.....--)))))).))))))))((((...))))......---.........)))........................--- ( -14.40, z-score = -1.23, R) >droYak2.chr2L 11045812 104 + 22324452 --GUUAUUCGUGCGCAAUUACAUAC-AUUAAUAUGAUUAA--CAUAUUUUUGCGCAUUGUCCUCGACAUACCAG---CAUAAAUAAGAAGCCCAGCCAGUUCCCAACUCCCG---- --....(((((((((((........-...((((((.....--)))))).))))))))((((...))))......---.........))).......................---- ( -14.40, z-score = -1.04, R) >droEre2.scaffold_4929 17316755 104 + 26641161 --GUUAUUCGUGCGCAAUUACAUAC-AUUAAUAUGAUUAA--CAUAUUUUUGCGCAUUGUCCUCGACAUACCAG---CAUAAAUAAAAAGCCCAGCCAGUUCCCAACUCCCG---- --(((....((((((((........-...((((((.....--)))))).))))))))((((...))))....))---)..................................---- ( -13.90, z-score = -1.22, R) >droAna3.scaffold_12916 5491171 110 + 16180835 --GUUAUUCUUGCGCAAUUACAUAC-AUUAAUAUGAUUAA--CAUAUUGUUGCGCAUUGUCCUUGACAUACCAUA-CCAUAACUUUAGAGCCCAGCCAGAUCCUGAUCCCCCCCAG --(((((...((((((((.......-...((((((.....--)))))))))))))).((((...)))).......-..))))).........(((.......)))........... ( -15.80, z-score = -1.59, R) >dp4.chr4_group1 1175713 104 - 5278887 --GUUAUUCGCGCGCAAUUACAUACGAUUAAUAUGAUUAAAACAUAUUUUUGCGCAAUGUCCUCGACACACGUG------AA-CCACAAACCAAUCCAGGAACCCACUUCCCG--- --((..(((((((((((............((((((.......)))))).))))))..((((...))))...)))------))-..))...........((((.....))))..--- ( -19.62, z-score = -2.62, R) >droPer1.super_5 1161638 105 + 6813705 --GUUAUCCGCGCGCAAUUACAUACGAUUAAUAUGAUUAAAACAUAUUUUUGCGCAAUGUCCUCGACACACGUG------AAACCACAAACCAAUCCAGGAACCCACUUCCCG--- --....(((..((((((............((((((.......)))))).))))))..((((...))))...(((------....)))...........)))............--- ( -16.82, z-score = -1.68, R) >droWil1.scaffold_181132 297468 109 + 1035393 AUUCUAUA-UUGCGCAAUUACAUAC-AUUAAUAUGAUU-AA-CAUAUUUUUGCGCAUUGUCCUCGACAUACAACAAGAAGAACCACAAUAUUAAAGCAGAUCCCCCUCCACCC--- .((((...-.(((((((........-...((((((...-..-)))))).))))))).((((...)))).......))))..................................--- ( -14.10, z-score = -2.37, R) >droVir3.scaffold_12723 3002733 94 + 5802038 --GUUAAU-GUGCGCAAUUGCAUAC-CUUAAUAUGAUU-AA-CAUAUUUUAGCGUACUU--GCCGCCA----GCCGAACCGACAACAAAACCACGCGCACUCCCUA---------- --......-((((((....((((((-((.((((((...-..-))))))..)).)))..)--))((...----..))..................))))))......---------- ( -17.90, z-score = -1.58, R) >droMoj3.scaffold_6500 25485589 92 - 32352404 --GUUAAU-GUGCGCAAUUGCAUAC-CUUAAUAUGAUUUAA-CAUAUUUU-GCGUACUUUUGCCACCA----AC--AAGCAACAACAAAACCACG--CACUCCCUA---------- --......-(((((........(((-(..((((((......-))))))..-).)))...((((.....----..--..))))...........))--)))......---------- ( -14.00, z-score = -1.25, R) >droGri2.scaffold_15126 7918056 92 - 8399593 --GUUAAU-GUGCGCAAUUGCAUAC-AUUAAUAUGAUU-AA-CAUAUUUUUGCGCACUCA-CCUGCCA----AC-AUACCAAGAACAGAACCACG--CACUCCCGA---------- --......-((((((((........-...((((((...-..-)))))).))))))))...-.......----..-....................--.........---------- ( -14.00, z-score = -0.97, R) >consensus __GUUAUUCGUGCGCAAUUACAUAC_AUUAAUAUGAUUAA__CAUAUUUUUGCGCAUUGUCCUCGACAUACCAG___CAUAAAUAAAAAACCAAGCCAGUUCCCAACUCCCC____ ..........(((((((............((((((.......)))))).)))))))............................................................ ( -8.53 = -8.73 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:25:53 2011