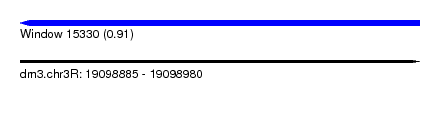
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,098,885 – 19,098,980 |
| Length | 95 |
| Max. P | 0.905617 |

| Location | 19,098,885 – 19,098,980 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 60.59 |
| Shannon entropy | 0.78612 |
| G+C content | 0.55638 |
| Mean single sequence MFE | -26.54 |
| Consensus MFE | -9.41 |
| Energy contribution | -9.99 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19098885 95 - 27905053 ------UCUGGCCCCGGAAACGCUUUCAAACCCACUUCUCCCGUCCGUCGACCACCA--UUGCCACAUCAUG---UGUAUGAGUGUAUCCGCUAAAAUGCAGGCCA ------...((((..(((.((((((.................(((....))).....--....(((.....)---))...)))))).)))((......)).)))). ( -23.20, z-score = -1.59, R) >droSim1.chr3R 18914470 97 - 27517382 ------UCUGGCCCCGGAAACGCUUUCAAACCCACUUCCCCCAUCCGUCGACCACCACCAUGCCACAUCAUG---UGUAUGAGUGUAUCCGCUAAAAUGCAGGCCA ------...((((..(((.(((((...................((....)).......(((((.((.....)---))))))))))).)))((......)).)))). ( -22.50, z-score = -1.71, R) >droSec1.super_0 19463720 97 - 21120651 ------UCUGGCCCCGGAAACGCUUUCAAACCCACUUCCCCCAUCCAUCGACUACCACCAUGCCACAUCAUG---UGUGUGAGUGUAUCCGCUAAAAUGCAGGCCA ------...((((..(((.((((((..................((....))....(((((((......))))---.))).)))))).)))((......)).)))). ( -23.90, z-score = -2.07, R) >droYak2.chr3R 19974072 81 - 28832112 ------UCUGGCCCCGGAAACGCUUUUCAACCCACU--CCCCAUCC--------------CGUCACAUCAUG---UGUGUGAGUGUAUCCGCUAAAAUGCAGGCCA ------...((((..(((.((((((....((.(((.--........--------------...........)---)).)))))))).)))((......)).)))). ( -21.75, z-score = -2.18, R) >droEre2.scaffold_4820 1514735 92 + 10470090 ------UCUGGCCCCGGAAACGCUCUCCAAUCCACAUCCCCG--CCGUCGACCAGCA---UGCCACAGCAUG---UGUGUGAGUGUAUCCGCUGAAAUGCAGGCCA ------...((((..(((.((((((.((..((.((.......--..)).))...(((---(((....)))))---)).).)))))).)))((......)).)))). ( -29.60, z-score = -1.70, R) >droWil1.scaffold_181089 11649823 106 + 12369635 CCCCCCCACAGGCUGCCAGGCUAUGUAUAGCCCCCCCUCCUCCGGCAUCUGGUUACCAUCUCCGGCCGGGAGAAUUGCAUAUGUGUGUGCACUUAAAUGCAGGCCA ..........((((((..(((((....))))).....((..(((((....(((....)))....)))))..))..((((((....)))))).......))).))). ( -32.80, z-score = 0.28, R) >droGri2.scaffold_14906 9886705 100 + 14172833 --CCAGCUCAGUCCUGCUCUUGGUCCCAGUUCCCAGCACCGCGGCUAUUCUGUAUCUGUGUGUGUGUGUGUGUGCUGCACA----UAUGCACUAGUAUGCAGUCCA --..((((..((..((((...((........)).))))..))))))...(((((((((.((((((((((((.....)))))----)))))))))).)))))).... ( -32.00, z-score = -1.09, R) >consensus ______UCUGGCCCCGGAAACGCUUUCAAACCCACUUCCCCCAUCCAUCGACCACCA___UGCCACAUCAUG___UGUAUGAGUGUAUCCGCUAAAAUGCAGGCCA .........((((.((....)).....................................................(((((.((((....))))...))))))))). ( -9.41 = -9.99 + 0.58)

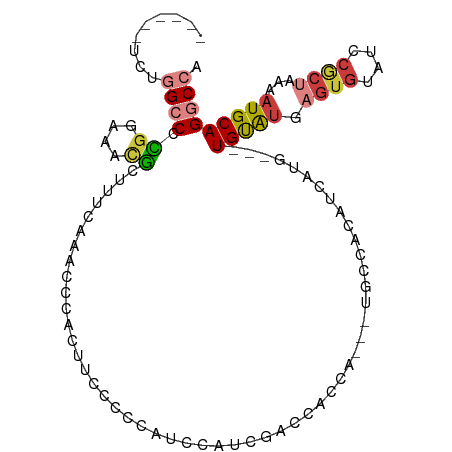
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:37:29 2011