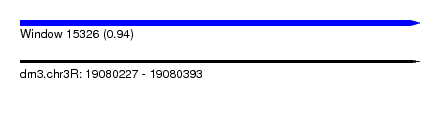
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,080,227 – 19,080,393 |
| Length | 166 |
| Max. P | 0.939686 |

| Location | 19,080,227 – 19,080,393 |
|---|---|
| Length | 166 |
| Sequences | 6 |
| Columns | 168 |
| Reading direction | forward |
| Mean pairwise identity | 70.80 |
| Shannon entropy | 0.56195 |
| G+C content | 0.46883 |
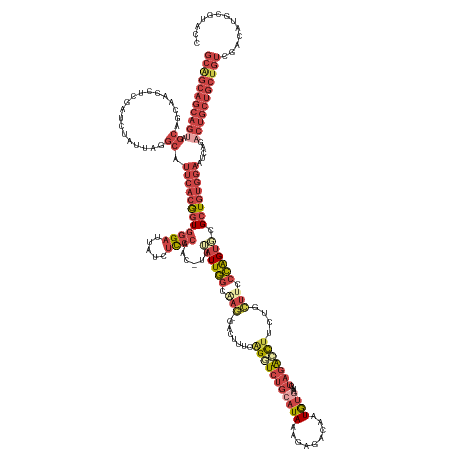
| Mean single sequence MFE | -51.62 |
| Consensus MFE | -23.93 |
| Energy contribution | -28.33 |
| Covariance contribution | 4.39 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.939686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
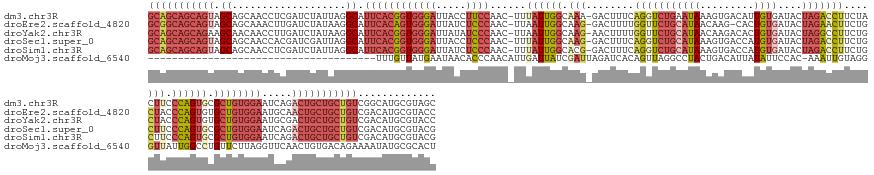
>dm3.chr3R 19080227 166 + 27905053 GCAGCAGCAGUAGCAGCAACCUCGAUCUAUUAGGCAUUCACGGUGGGAUUACCUUCCAAC-UUUAUUGGCAAA-GACUUUCAGGUCUGAAUAAAGUGACAUUGUGAUACUAGACCUUCUACUUCCCAGUGCGCUGUGGAAUCAGACUGCUGCUGUCGGCAUGCGUAGC (((((.((((((((((.............((((....(((((((((((......))).((-((((((.....(-((((....))))).))))))))..))))))))..))))....(((..(((((((....))).))))..)))))))))))))..)).)))..... ( -56.30, z-score = -2.08, R) >droEre2.scaffold_4820 1495851 165 - 10470090 GCGGCAGCAGUAGCAGCAAACUUGAUCUAUAAGGCAUUCACAGUGGGAUUAUCUCCCAAC-UUAAUUGGCAAG-GACUUUUGGUUCUGCAUAACAAG-CACUGUGAUACUAGAACUUCUGCUACCCAGUGUGCUGUGGAAUGCAACUGCUGCUGUCGACAUGCGUACC .((((((((((((.......((((.....))))(((((((((((((((.....))))...-.......(((.(-(((.....))))))).......(-(((((.(.((.(((.....))).)).)))))))))))).))))))..))))))))))))........... ( -56.80, z-score = -2.23, R) >droYak2.chr3R 19955049 166 + 28832112 GCAGCAGCAGAAGCAACAACCUUGAUCUAUAAGGCAUUCACGGUGGGAUUAUAUCCCAAC-UUAAUUGGCAAG-AACUUUUGGUUCUGCAUAACAAGACACUGUGAUACUAGGCCUUCUGCUACCCAGUGUGCUGUGGAAUGCGACUGCUGCUGUCGACAUGCGUACC ((((((((((..(((...............(((((..((((((((((((...)))))..(-((.....(((.(-(((.....))))))).....)))..)))))))......)))))......(((((....))).))..)))..))))))))))............. ( -56.60, z-score = -2.78, R) >droSec1.super_0 19444823 166 + 21120651 GCAGCAGCAGUAGCAGCAACCACGAUCGAUUAGGCAUUCACGGUGGGAUUACCUCCCAAC-UUUAUUGGCAAG-GACUUUCAGGUCUGCAUAAAGUGACCAUGUGAUACUAGACCUUCUGCUUCCCAGUGCGCUGUGGAAUCAGACUGCUGCUGUCGACAUGCGUACG (((.(.((((((((((.............((((....(((((((((((.....)))).((-(((((..(((..-((((....)))))))))))))).)))..))))..))))....((((.(((((((....))).)))).)))))))))))))).)...)))..... ( -56.80, z-score = -1.77, R) >droSim1.chr3R 18895666 166 + 27517382 GCAGCAGCAGUAGCAGCAACCUCGAUCUAUUAGGCAUUCACGGUGGGAUUAUCUCCCAAC-UUUAUUGGCACG-GACUUUCAGGUCUGCAUAAAGUGACCAUGUGAUACUAGACCUUCUGCUUCCCAGUGCGCUGUGGAAUCAGACUGCUGCUGUCGACAUGCGUACG (((.(.((((((((((.............((((....(((((((((((.....)))).((-(((((..(((..-((((....)))))))))))))).)))..))))..))))....((((.(((((((....))).)))).)))))))))))))).)...)))..... ( -56.80, z-score = -1.92, R) >droMoj3.scaffold_6540 10619916 129 + 34148556 --------------------------------------UUUGUUAUGAAUAACACCCAACAUUGAUUAUCGAUUAGAUCACAGUUAGGCCUACUGACAUUAUAUUCCAC-AAAUUGUAGGGUUAUUGGCCUGUUCUUAGGUUCAACUGUGACAGAAAAUAUGCGCACU --------------------------------------..(((((....)))))......((((.....))))....((((((((..(((((..((((..(((..((((-.....)).))..))).....))))..)))))..))))))))................. ( -26.40, z-score = -0.32, R) >consensus GCAGCAGCAGUAGCAGCAACCUCGAUCUAUUAGGCAUUCACGGUGGGAUUAUCUCCCAAC_UUUAUUGGCAAG_GACUUUCAGGUCUGCAUAAAGAGACAAUGUGAUACUAGACCUUCUGCUUCCCAGUGCGCUGUGGAAUCAGACUGCUGCUGUCGACAUGCGUACC (((((((((((.((...................)).((((((((((((.....))))......((((((.(((........(((((((((((.........))))....)))))))....))).)))))).)))))))).....)))))))))))............. (-23.93 = -28.33 + 4.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:37:25 2011