| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,071,994 – 19,072,088 |
| Length | 94 |
| Max. P | 0.939862 |

| Location | 19,071,994 – 19,072,088 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 69.07 |
| Shannon entropy | 0.64881 |
| G+C content | 0.52982 |
| Mean single sequence MFE | -25.24 |
| Consensus MFE | -10.11 |
| Energy contribution | -10.84 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.939862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19071994 94 - 27905053 -------AACGCCCA-ACGC-CCACCACUGGGCCAC--UGUUCA--CAUUUGAUGUGCAUUUGCAUU-CGUGGCCCUCCGGCUAUCGAAUGCUUUUUAGCCCACGACA -------........-....-.......((((((((--...(((--....))).))).....(((((-(((((((....))))).)))))))......)))))..... ( -27.70, z-score = -2.11, R) >droSim1.chr3R 18887556 94 - 27517382 -------ACCGCCCA-CCGC-CCACCACUGGGCCAC--UGUUCA--CAUUUGAUGUGCAUUUGCAUU-CGUGGCCCUCCGGCUAUCGAAUGCUUUUUAGCCCACGACA -------........-....-.......((((((((--...(((--....))).))).....(((((-(((((((....))))).)))))))......)))))..... ( -27.70, z-score = -2.20, R) >droSec1.super_0 19436695 94 - 21120651 -------ACCGUCCA-CCUC-CCACCACUGGGCCAC--UGUUCA--CAUUUGAUGUGCAUUUGCAUU-CGUGGCCCUCCGGCUAUCGAAUGCUUUUUAGCCCACGACA -------........-....-.......((((((((--...(((--....))).))).....(((((-(((((((....))))).)))))))......)))))..... ( -27.70, z-score = -2.81, R) >droYak2.chr3R 19946643 94 - 28832112 -------ACCACCCA-CCAC-CCACCUCUGGGCCAC--UGUUCA--CAUUUGAUGUGCAUUUGCAUU-CGUGGCCCUCCGGCUAUCGAAUGUUUUUUAGCCCACGACA -------........-....-.....((((((((((--...(((--....))).))).....(((((-(((((((....))))).)))))))......))))).)).. ( -26.10, z-score = -2.63, R) >droEre2.scaffold_4820 1485982 92 + 10470090 ---------CAUGCA-CCGC-CCACCUCUGGGCCAC--UGUUCA--CAUUUGAUGUGCAUUUGCAUU-CGUGGCCCUCCGGCUAUCGAAUGCUUUUUAGCCCACGACA ---------.(((((-(.((-(((....)))))...--...(((--....))).))))))..(((((-(((((((....))))).)))))))................ ( -31.20, z-score = -3.28, R) >droAna3.scaffold_13340 15378088 102 + 23697760 ---GCCACCCAUCCACCCAU-CCACCCAUGUGCCACCUCAUCCG--GAUUUGAUGUGCAUUUGCAUUCCGUGGCCCUAAGGCUAUCGAAUGCUUUUUAGCCCACGACA ---((...............-........((((.((.(((....--....))).))))))..((((((.((((((....)))))).))))))......))........ ( -23.30, z-score = -1.38, R) >dp4.chr2 21215675 94 + 30794189 ----------CCCAACCCGC-GCCCACAUGGGUGAUCCCCUUCA--AAUUUGAUGUGCAUUUGCAUU-CGUGGCCCUCAGGCUAUCGAAUGCUUUUUAGUCCACGACA ----------.........(-((((....)))))..........--....((.((((.(((.(((((-(((((((....))))).))))))).....))).)))).)) ( -27.90, z-score = -2.31, R) >droPer1.super_0 10901492 94 - 11822988 ----------CCCAACCCGC-GCCCACAUGGGUGAUCCCCUUCA--CAUUUGAUGUGCAUUUGCAUU-CGUGGCCCUCAGGCUAUCGAAUGCUUUUUAGUCCACGACA ----------........((-((((....))((((......)))--).......))))....(((((-(((((((....))))).)))))))......(((...))). ( -28.60, z-score = -2.52, R) >droWil1.scaffold_181130 8682577 108 - 16660200 AGUAUCCGGCUCCAAGUCAUGUUGCAAACUCCCUGUUCUAGUCAUCUAACUGAUGUGCAUUUGCAUUUUCCAUCCCUGAGGCUAUCGAAUGUUUUUUAGUUCACGACA .(((...(((.....)))....)))...............(((....((((((...(((((((.((...((........))..)))))))))...))))))...))). ( -17.30, z-score = 0.67, R) >droVir3.scaffold_13047 8057102 72 - 19223366 -----------------CACUCCCCAUUUAUGCCCCAAGGCUAU--CGAAUGCAGCGCAUUGUCAU--UGUUGCC-----AGCGCCGGCUGCUCCUUU---------- -----------------..............(((....)))...--.....(((((((.(((.((.--...)).)-----)).))..)))))......---------- ( -14.90, z-score = 0.76, R) >consensus _________CACCCA_CCGC_CCACCACUGGGCCAC__UGUUCA__CAUUUGAUGUGCAUUUGCAUU_CGUGGCCCUCCGGCUAUCGAAUGCUUUUUAGCCCACGACA .................................................((((...(((((((......((((((....)))))))))))))...))))......... (-10.11 = -10.84 + 0.73)


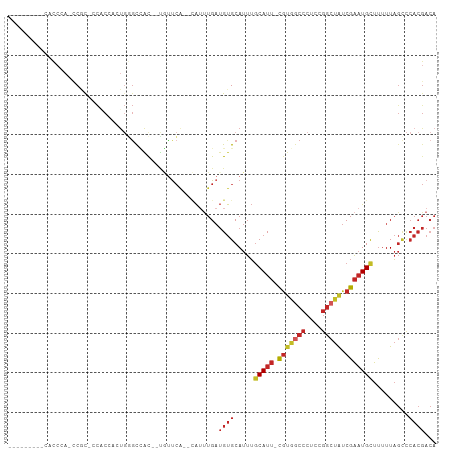
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:37:23 2011