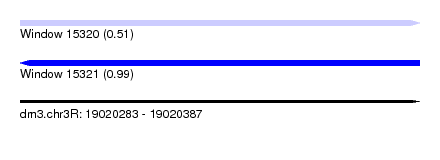
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,020,283 – 19,020,387 |
| Length | 104 |
| Max. P | 0.989622 |

| Location | 19,020,283 – 19,020,387 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 85.29 |
| Shannon entropy | 0.26907 |
| G+C content | 0.43580 |
| Mean single sequence MFE | -25.82 |
| Consensus MFE | -20.06 |
| Energy contribution | -21.50 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.507905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
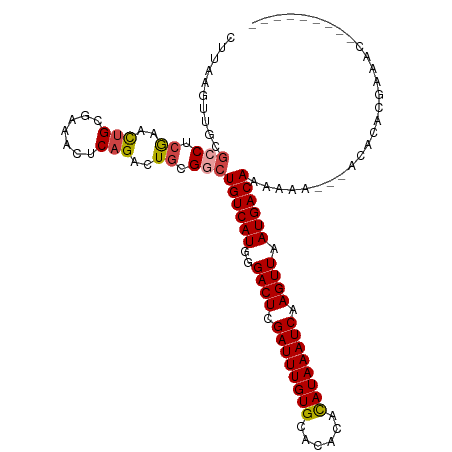
>dm3.chr3R 19020283 104 + 27905053 ---------GUUUCGUGUUUUU-UUUUUUGUCAUUAACUUGAUUUAUGUGUGUGCACAAAUCGAGUCCCAUGACAGCCGCAGUCUGAGUUUCGCAGUUCGAGUCGCAACUUAAG ---------.............-....(((((((..((((((((..((((....))))))))))))...)))))))........((((((.(((.......).)).)))))).. ( -22.00, z-score = -0.52, R) >droEre2.scaffold_4820 1425531 101 - 10470090 ---------GUUUCGUGUGU----UUUUUGUCAUUAACUUGAUUUAUGUGUGUGCACAAAUCGAGUCCCAUGACAGCCGCAGUCUGAGUUUCGCAGUUCGAGGCGCAACUUAAG ---------.....(((.((----....((((((..((((((((..((((....))))))))))))...)))))))))))....((((((.(((........))).)))))).. ( -28.50, z-score = -1.76, R) >droYak2.chr3R 19890403 114 + 28832112 GUUUCGUGUGUUUUUUUUUUCAGUUUUUUGUCAUUAACUUGAUUUAUGUGUGUGCACAAAUCGAGUCCCAUGACAGCCGCAGUCUGAGUUUCGCAGUUCGCGGCGCAACUUAAG ......(((...................((((((..((((((((..((((....))))))))))))...))))))(((((...(((.......)))...))))))))....... ( -31.00, z-score = -2.38, R) >droSec1.super_0 19383525 102 + 21120651 ---------GUUUCGUGUGU---UUUUUUGUCAUUAACUUGAUUUAUACGUGUGCACAAAUCGAGUCCCAUGACAGCCGCAGUCUGAGUUUCGCAGUUCGAGGCGCAACUUAAG ---------.....(((.((---.....((((((..(((((((((....((....)))))))))))...)))))))))))....((((((.(((........))).)))))).. ( -23.50, z-score = -0.32, R) >droSim1.chr3R 18832932 102 + 27517382 ---------GUUUCGUGUGU---UUUUUUGUCAUUAACUUGAUUUAUGUGUGUGCACAAAUCGAGUCCCAUGACAGCCGCAGUCUGAGUUUCGCAGUUCGAGGCGCAACUUAAG ---------.....(((.((---.....((((((..((((((((..((((....))))))))))))...)))))))))))....((((((.(((........))).)))))).. ( -28.10, z-score = -1.65, R) >droAna3.scaffold_13340 15322753 84 - 23697760 -------------CGUGUGU---UUUUUUGUCAUUAACUUGAUUUAUGUGUGUGCACAAAUCGAGUCCCAUGACAGCCCGAGUCGGAG--UCGCAACUUGAG------------ -------------..((((.---.....((((((..((((((((..((((....))))))))))))...))))))(((((...))).)--))))).......------------ ( -21.80, z-score = -1.35, R) >consensus _________GUUUCGUGUGU___UUUUUUGUCAUUAACUUGAUUUAUGUGUGUGCACAAAUCGAGUCCCAUGACAGCCGCAGUCUGAGUUUCGCAGUUCGAGGCGCAACUUAAG ...........................(((((((..(((((((((.(((....))).)))))))))...)))))))........((((((.(((........))).)))))).. (-20.06 = -21.50 + 1.44)


| Location | 19,020,283 – 19,020,387 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 85.29 |
| Shannon entropy | 0.26907 |
| G+C content | 0.43580 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -20.61 |
| Energy contribution | -21.03 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.989622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19020283 104 - 27905053 CUUAAGUUGCGACUCGAACUGCGAAACUCAGACUGCGGCUGUCAUGGGACUCGAUUUGUGCACACACAUAAAUCAAGUUAAUGACAAAAAA-AAAAACACGAAAC--------- .............(((..(((((..........))))).((((((..((((.((((((((......)))))))).)))).)))))).....-.......)))...--------- ( -23.80, z-score = -2.13, R) >droEre2.scaffold_4820 1425531 101 + 10470090 CUUAAGUUGCGCCUCGAACUGCGAAACUCAGACUGCGGCUGUCAUGGGACUCGAUUUGUGCACACACAUAAAUCAAGUUAAUGACAAAAA----ACACACGAAAC--------- .....(((..(((.((..(((.......)))..)).)))((((((..((((.((((((((......)))))))).)))).))))))...)----)).........--------- ( -25.70, z-score = -2.25, R) >droYak2.chr3R 19890403 114 - 28832112 CUUAAGUUGCGCCGCGAACUGCGAAACUCAGACUGCGGCUGUCAUGGGACUCGAUUUGUGCACACACAUAAAUCAAGUUAAUGACAAAAAACUGAAAAAAAAAACACACGAAAC ....((((..((((((..(((.......)))..))))))((((((..((((.((((((((......)))))))).)))).))))))...))))..................... ( -32.20, z-score = -4.16, R) >droSec1.super_0 19383525 102 - 21120651 CUUAAGUUGCGCCUCGAACUGCGAAACUCAGACUGCGGCUGUCAUGGGACUCGAUUUGUGCACACGUAUAAAUCAAGUUAAUGACAAAAAA---ACACACGAAAC--------- .....(((..(((.((..(((.......)))..)).)))((((((..((((.(((((((((....))))))))).)))).))))))....)---)).........--------- ( -27.30, z-score = -2.46, R) >droSim1.chr3R 18832932 102 - 27517382 CUUAAGUUGCGCCUCGAACUGCGAAACUCAGACUGCGGCUGUCAUGGGACUCGAUUUGUGCACACACAUAAAUCAAGUUAAUGACAAAAAA---ACACACGAAAC--------- .....(((..(((.((..(((.......)))..)).)))((((((..((((.((((((((......)))))))).)))).))))))....)---)).........--------- ( -25.70, z-score = -2.28, R) >droAna3.scaffold_13340 15322753 84 + 23697760 ------------CUCAAGUUGCGA--CUCCGACUCGGGCUGUCAUGGGACUCGAUUUGUGCACACACAUAAAUCAAGUUAAUGACAAAAAA---ACACACG------------- ------------(((.(((((...--...))))).))).((((((..((((.((((((((......)))))))).)))).)))))).....---.......------------- ( -23.60, z-score = -3.16, R) >consensus CUUAAGUUGCGCCUCGAACUGCGAAACUCAGACUGCGGCUGUCAUGGGACUCGAUUUGUGCACACACAUAAAUCAAGUUAAUGACAAAAAA___ACACACGAAAC_________ ..........(((.((..(((.......)))..)).)))((((((..((((.((((((((......)))))))).)))).))))))............................ (-20.61 = -21.03 + 0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:37:21 2011