| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,997,388 – 18,997,481 |
| Length | 93 |
| Max. P | 0.997719 |

| Location | 18,997,388 – 18,997,481 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 68.23 |
| Shannon entropy | 0.57885 |
| G+C content | 0.42503 |
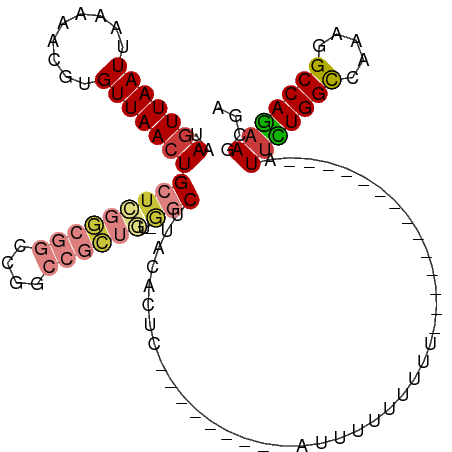
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -17.11 |
| Energy contribution | -18.49 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.997719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
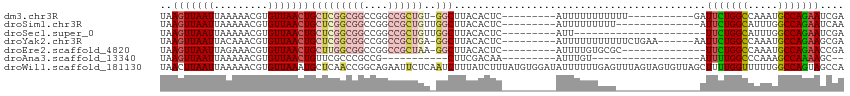
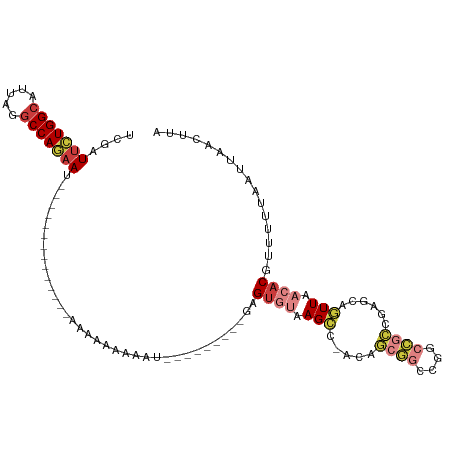
>dm3.chr3R 18997388 93 + 27905053 UAAGUUAAUUAAAAACGUGUUAACUGCUCGGCGGCCGGCCGCUGU-GGCUUACACUC---------AUUUUUUUUUUU-----------GAUUCUGGCCAAAUGCCAGAAUCGA ................((((.....(((((((((....)))))).-)))..))))..---------..........((-----------(((((((((.....))))))))))) ( -32.90, z-score = -4.01, R) >droSim1.chr3R 18809674 91 + 27517382 UAAGUUAAUUAAAAACGUGUUAACUGCUCGGCGGCCGGCCGCUGUUGGCUUACACUC---------AUUUUUUUUU--------------AUUCUGGCAUUUGGCCAGAAUCAA ................((((.....(((((((((....))))))..)))..))))..---------..........--------------((((((((.....))))))))... ( -27.50, z-score = -2.66, R) >droSec1.super_0 19361206 83 + 21120651 UAAGUUAAUUAAAAACGUGUUAACUGCUCGGCGGCCGGCCGCUGUUGGCUUACACUC---------AUU----------------------UUCUGGCAUUUGGCCAGAAUCGA ................((((.....(((((((((....))))))..)))..))))..---------...----------------------(((((((.....))))))).... ( -26.90, z-score = -2.21, R) >droYak2.chr3R 19866102 98 + 28832112 UAAGUUAAUUACAAACGUGUUAACUGCUCGGCGGCCGGCCGCUGA-GGCUUACACUC---------AUUUUUUUUUUUCUGAA------AAUUCUGGCCAAAUGCCAGAAGCGA ..(((((((.((....))))))))).((((((((....)))))))-)(((.......---------((((((........)))------)))((((((.....))))))))).. ( -31.70, z-score = -2.86, R) >droEre2.scaffold_4820 1402733 90 - 10470090 UAAGUUAAUUAGAAACGUGUUAACUGCUUGGCGGCCGGCCGCUAA-GGCUUACACUC---------AUUUUGUGCGC--------------UUCUGGCCAAAUGCCAGAACCGA ..(((((((.........))))))).((((((((....)))))))-).....(((..---------.....)))((.--------------(((((((.....))))))).)). ( -30.50, z-score = -2.04, R) >droAna3.scaffold_13340 15298222 74 - 23697760 UAAGUUAAUUAAAAACGUGUUAACUGUUCGCCCGCCG-----------CUUCGACAA---------AUUUGU------------------AUUUUGGCCCAAAGCCAAAAGC-- ..(((((((.........)))))))(((.((.....)-----------)...)))..---------......------------------.(((((((.....)))))))..-- ( -11.70, z-score = -0.09, R) >droWil1.scaffold_181130 8596566 114 + 16660200 UAACUUAAUUAAAAACGUGUUAAAUGCUCAACCGGCAGAAUUCUCAAUCUUUAUCUUUAUGUGGAUAUUUUUUGAGUUUAGUAGUGUUAGCUUUUGGUUUUUGGCCAGUAGCCA ((((...((((((...........((((.....)))).....(((((....(((((......)))))....)))))))))))...))))(((.(((((.....))))).))).. ( -22.90, z-score = -0.78, R) >consensus UAAGUUAAUUAAAAACGUGUUAACUGCUCGGCGGCCGGCCGCUGU_GGCUUACACUC_________AUUUUUUUUU______________AUUCUGGCCAAAGGCCAGAAGCGA ..(((((((.........)))))))(((((((((....))))))..)))..........................................(((((((.....))))))).... (-17.11 = -18.49 + 1.37)
| Location | 18,997,388 – 18,997,481 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 68.23 |
| Shannon entropy | 0.57885 |
| G+C content | 0.42503 |
| Mean single sequence MFE | -22.99 |
| Consensus MFE | -13.63 |
| Energy contribution | -14.96 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18997388 93 - 27905053 UCGAUUCUGGCAUUUGGCCAGAAUC-----------AAAAAAAAAAAU---------GAGUGUAAGCC-ACAGCGGCCGGCCGCCGAGCAGUUAACACGUUUUUAAUUAACUUA ..(((((((((.....)))))))))-----------......((((((---------(.((....((.-.(.((((....)))).).)).....)).))))))).......... ( -28.50, z-score = -2.92, R) >droSim1.chr3R 18809674 91 - 27517382 UUGAUUCUGGCCAAAUGCCAGAAU--------------AAAAAAAAAU---------GAGUGUAAGCCAACAGCGGCCGGCCGCCGAGCAGUUAACACGUUUUUAAUUAACUUA ...((((((((.....))))))))--------------...(((((..---------..((((.(((...(.((((....)))).)....))).)))).))))).......... ( -24.10, z-score = -2.04, R) >droSec1.super_0 19361206 83 - 21120651 UCGAUUCUGGCCAAAUGCCAGAA----------------------AAU---------GAGUGUAAGCCAACAGCGGCCGGCCGCCGAGCAGUUAACACGUUUUUAAUUAACUUA ....(((((((.....)))))))----------------------...---------..((((.(((...(.((((....)))).)....))).))))................ ( -22.60, z-score = -1.46, R) >droYak2.chr3R 19866102 98 - 28832112 UCGCUUCUGGCAUUUGGCCAGAAUU------UUCAGAAAAAAAAAAAU---------GAGUGUAAGCC-UCAGCGGCCGGCCGCCGAGCAGUUAACACGUUUGUAAUUAACUUA ....(((((((.....)))))))..------.................---------..((((.((((-((.((((....)))).)))..))).))))................ ( -27.20, z-score = -1.38, R) >droEre2.scaffold_4820 1402733 90 + 10470090 UCGGUUCUGGCAUUUGGCCAGAA--------------GCGCACAAAAU---------GAGUGUAAGCC-UUAGCGGCCGGCCGCCAAGCAGUUAACACGUUUCUAAUUAACUUA .((.(((((((.....)))))))--------------.))........---------..((((.((((-((.((((....)))).)))..))).))))................ ( -28.70, z-score = -1.56, R) >droAna3.scaffold_13340 15298222 74 + 23697760 --GCUUUUGGCUUUGGGCCAAAAU------------------ACAAAU---------UUGUCGAAG-----------CGGCGGGCGAACAGUUAACACGUUUUUAAUUAACUUA --..(((((((.....))))))).------------------......---------.....((((-----------((...(((.....)))....))))))........... ( -13.90, z-score = 0.16, R) >droWil1.scaffold_181130 8596566 114 - 16660200 UGGCUACUGGCCAAAAACCAAAAGCUAACACUACUAAACUCAAAAAAUAUCCACAUAAAGAUAAAGAUUGAGAAUUCUGCCGGUUGAGCAUUUAACACGUUUUUAAUUAAGUUA ..(((((((((...........((......))......(((((....((((........))))....)))))......))))))..)))......................... ( -15.90, z-score = -0.13, R) >consensus UCGAUUCUGGCAUUAGGCCAGAAU______________AAAAAAAAAU_________GAGUGUAAGCC_ACAGCGGCCGGCCGCCGAGCAGUUAACACGUUUUUAAUUAACUUA ....(((((((.....)))))))....................................((((.(((.....((((....))))......))).))))................ (-13.63 = -14.96 + 1.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:37:18 2011