
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,377,893 – 8,377,943 |
| Length | 50 |
| Max. P | 0.921654 |

| Location | 8,377,893 – 8,377,943 |
|---|---|
| Length | 50 |
| Sequences | 12 |
| Columns | 56 |
| Reading direction | reverse |
| Mean pairwise identity | 68.13 |
| Shannon entropy | 0.65565 |
| G+C content | 0.48024 |
| Mean single sequence MFE | -12.83 |
| Consensus MFE | -6.88 |
| Energy contribution | -6.66 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.71 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.921654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
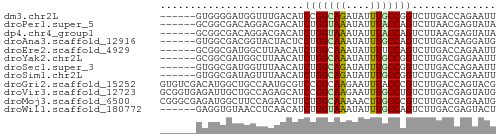
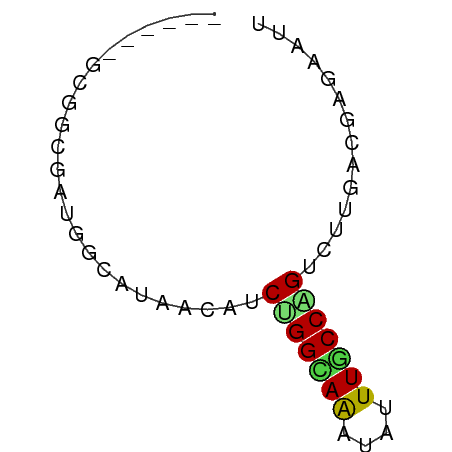
>dm3.chr2L 8377893 50 - 23011544 ------GUGGGGAUGGUUUGACAUCCGGCAGAUAUUUGCCGGUCUUGACCAGAAUU ------....(((((......)))))(((((....)))))(((....)))...... ( -16.80, z-score = -1.89, R) >droPer1.super_5 1132107 50 - 6813705 ------GCGGCGACAGGACGACAUCUGGUAAAUAUUUACCAGUCUUAACGAGUAUA ------((..((..((((((....)((((((....)))))))))))..)).))... ( -10.10, z-score = -1.04, R) >dp4.chr4_group1 1146307 50 + 5278887 ------GCGGCGACAGGACGACAUCUGGUAAAUAUUUACCAGUCUUAACGAGUAUA ------((..((..((((((....)((((((....)))))))))))..)).))... ( -10.10, z-score = -1.04, R) >droAna3.scaffold_12916 5462217 50 - 16180835 ------GUGGCGACGGUACUACUCCUGGCAAAUAUUUGCCAGUCUUGACAAGGAUG ------((((........))))..(((((((....)))))))((((....)))).. ( -12.70, z-score = -1.01, R) >droEre2.scaffold_4929 17287329 50 - 26641161 ------GCGGCGAUGGCUUAACAUCUGGCAAAUAUUUUCCAGUCUUGACCAGAAUU ------..(((....))).....((((((((.............))).)))))... ( -8.22, z-score = 0.14, R) >droYak2.chr2L 11015921 50 - 22324452 ------GCGGCGAUGGCUUAACAUCUGGCAAAUAUUUGCCGGUCUUGACGAGAAUU ------..(((((((......))))((((((....)))))))))............ ( -12.90, z-score = -1.16, R) >droSec1.super_3 3874492 50 - 7220098 ------GUGGCGAUGGUUUAACAUCUGGCAGAUAUUUGCCGGUCUUGACCAGAAUU ------.......(((((......(((((((....)))))))....)))))..... ( -14.30, z-score = -1.67, R) >droSim1.chr2L 8164480 50 - 22036055 ------GUGGCGAUAGUUUAACAUCUGGCAGAUAUUUGCCGGUCUUGACCAGAAUU ------..(((((..((((..(....)..))))..)))))(((....)))...... ( -10.90, z-score = -0.71, R) >droGri2.scaffold_15252 16844174 56 + 17193109 GUGUCGACAUGGCUGCCAAUGCGUCCGGCAAGAAUUUACCCGUCUUGACCAGUACG (((.(.....((.(((....))).))(((((((.........))))).)).)))). ( -11.10, z-score = 0.52, R) >droVir3.scaffold_12723 3476272 56 + 5802038 GCGGUGAGAUUGCUGCCAGAGCAUCCGGCAAGAAUUUGCCUGUCUUGACGAGUAUG .((..(((((((((.....))))...(((((....))))).)))))..))...... ( -14.10, z-score = -0.12, R) >droMoj3.scaffold_6500 9030077 56 - 32352404 CGGGCGAGAUGGCUUCCAGAGCUUCUGGCAAAAACUUGCCCGUCUUGACGAGAAUG ((((((((.......((((.....))))......))))))))((((...))))... ( -18.62, z-score = -1.58, R) >droWil1.scaffold_180772 726012 50 + 8906247 ------GAGGUGUAACCUCAACAUCUGGUAAAUAUUUGCCAGUCUUGACGAGUACU ------((((.....))))..((.(((((((....)))))))...))......... ( -14.10, z-score = -2.30, R) >consensus ______GCGGCGAUGGCAUAACAUCUGGCAAAUAUUUGCCAGUCUUGACGAGAAUU ........................(((((((....))))))).............. ( -6.88 = -6.66 + -0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:25:51 2011