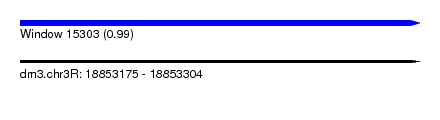
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,853,175 – 18,853,304 |
| Length | 129 |
| Max. P | 0.985588 |

| Location | 18,853,175 – 18,853,304 |
|---|---|
| Length | 129 |
| Sequences | 7 |
| Columns | 131 |
| Reading direction | forward |
| Mean pairwise identity | 74.81 |
| Shannon entropy | 0.48512 |
| G+C content | 0.56627 |
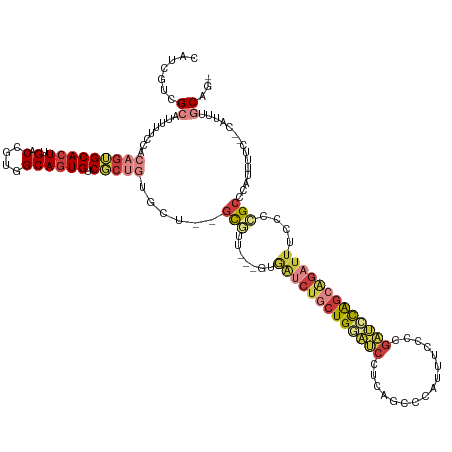
| Mean single sequence MFE | -35.37 |
| Consensus MFE | -19.79 |
| Energy contribution | -21.60 |
| Covariance contribution | 1.82 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.985588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
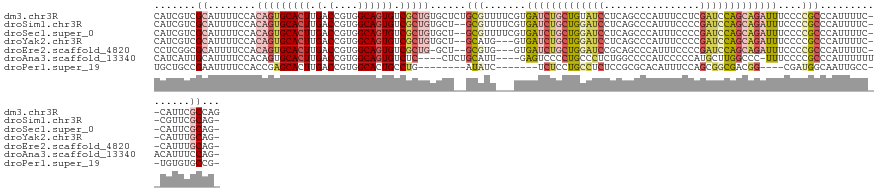
>dm3.chr3R 18853175 129 + 27905053 CAUCGUCGCAUUUUCCACAGUGCACUUGACCGUGGCAGUGUCGCUGUGCUCUGCGUUUUCGUGAUCUGCUGUAUCCUCAGCCCAUUUCCUCGAUCCAGCAGAUUUCCCCGCCCAUUUUC--CAUUCGCCAG .......((......(((((((((((.(.(....)))))).))))))).(((((....(((.((...((((......)))).....))..)))....))))).......))........--.......... ( -31.60, z-score = -2.01, R) >droSim1.chr3R 18667143 126 + 27517382 CAUCGUCGCAUUUUCCACAGUGCACUUGACCGUGGCAGUGUCGCUGUGCU--GCGUUUUCGUGAUCUGCUGGAUCCUCAGCCCAUUUCCCCGAUCCAGCAGAUUUCCCCGCCCAUUUUC--CGUUCGCAG- .......((......(((((((((((.(.(....)))))).)))))))..--(((.......(((((((((((((................)))))))))))))....)))........--.....))..- ( -39.29, z-score = -3.47, R) >droSec1.super_0 19216746 126 + 21120651 CAUCGUCGCAUUUUCCACAGUGCACUUGACCGUGGCAGUGUCGCUGUGCU--GCGUUUUCGUGAUCUGCUGGAUCCUCAGCCCAUUUCCCCGAUCCAGCAGAUUUCCCCGCCCAUUUUC--CAUUCGCAG- .......((......(((((((((((.(.(....)))))).)))))))..--(((.......(((((((((((((................)))))))))))))....)))........--.....))..- ( -39.29, z-score = -3.68, R) >droYak2.chr3R 19715250 123 + 28832112 CAUCGUCGCAUUUUCCACAGUGCACUUGACCGUGGCAGUGUCGCUGUGCU--GCAUG---GUGAUCUGCUGGAUCCUCAGCCCAUUUCCCCGAUCCAGCAGAUUUCCCCGCCCAUUUUC--CAUUUGCAG- .......(((.....(((((((((((.(.(....)))))).)))))))..--((..(---(.(((((((((((((................))))))))))))).))..))........--....)))..- ( -41.89, z-score = -3.69, R) >droEre2.scaffold_4820 1253608 122 - 10470090 CCUCGGCGCAUUUUCCACAGUGCACUUGACCGUGGCAGUGUCGCUG-GCU--GCGUG---GUGAUCUGCUGGAUCCGCAGCCCAUUUCCCCGAUCCAGCAGAUUUCCCCGCCCAUUUUC--CAUUUGCAG- ..((((((((((..((((.((.......)).)))).)))).)))))-)..--(((.(---(.(((((((((((((................))))))))))))).)).)))........--.........- ( -43.79, z-score = -2.73, R) >droAna3.scaffold_13340 11231695 121 - 23697760 CAUCAUUGCAUUUUCCACAGUGCACUUGACCGUGGCAGUGUCUC----CUCUGCAUU----GAGUCCCCUGCCCUCUGGCCCCAUCCCCCAUGCUUGGCCC-UUUCCCCGCCCAUUUUUUACAUUUCCAG- ..(((.((((((......))))))..)))..(.(((((.(....----(((......----)))...)))))))...((((.(((.....)))...)))).-............................- ( -21.90, z-score = -0.61, R) >droPer1.super_19 109230 109 - 1869541 UGCUGCCGAAUUUUCCACCGAGCACUUGACCGUGGCACUGCCUG--------AUAUC-------UCUCCUGCCUCUCCGCGCACAUUUCCAGCGGCGACGG----CGAUGGCAAUUGCC--UGUGUGCCG- ...(((((..........)).))).........(((((......--------.....-------.....((((((.((((...........)))).)).))----))..(((....)))--...))))).- ( -29.80, z-score = 0.52, R) >consensus CAUCGUCGCAUUUUCCACAGUGCACUUGACCGUGGCAGUGUCGCUGUGCU__GCGUU___GUGAUCUGCUGGAUCCUCAGCCCAUUUCCCCGAUCCAGCAGAUUUCCCCGCCCAUUUUC__CAUUUGCAG_ .......((........(((((((((.(.(....)))))).)))))......(((.......(((((((((((((................)))))))))))))....)))...............))... (-19.79 = -21.60 + 1.82)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:37:05 2011