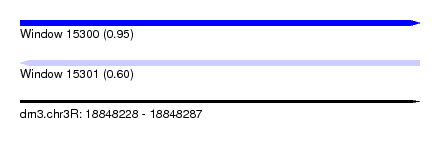
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,848,228 – 18,848,287 |
| Length | 59 |
| Max. P | 0.953134 |

| Location | 18,848,228 – 18,848,287 |
|---|---|
| Length | 59 |
| Sequences | 6 |
| Columns | 66 |
| Reading direction | forward |
| Mean pairwise identity | 76.19 |
| Shannon entropy | 0.43090 |
| G+C content | 0.48603 |
| Mean single sequence MFE | -17.60 |
| Consensus MFE | -12.70 |
| Energy contribution | -14.03 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.953134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
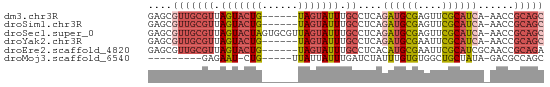
>dm3.chr3R 18848228 59 + 27905053 GAGCGUUGCGUUAGUACUG------UAGUAUUUGCCUCAGAUGCGAGUUCGCAUCA-AACCGCAGC ....(((((((.(((((..------..))))).))....((((((....)))))).-....))))) ( -19.60, z-score = -2.28, R) >droSim1.chr3R 18662300 59 + 27517382 GAGCGUUGCGUUAGUACUG------UAGUAUUUGCCUCAGAUGCGAGUUCGCAUCA-AACCGCAGC ....(((((((.(((((..------..))))).))....((((((....)))))).-....))))) ( -19.60, z-score = -2.28, R) >droSec1.super_0 19211881 65 + 21120651 GAGCGUUGCGUUAGUACUAGUGCGUUAGUAUUUGCCUCAGAUGCGAGUUCGCAUCA-AACCGCAGC ....(((((((.((((((((....)))))))).))....((((((....)))))).-....))))) ( -22.30, z-score = -2.36, R) >droYak2.chr3R 19710255 59 + 28832112 GAGCGUUGCGUUAGUACUG------UAGUAUUUGCCUCAGAUGCGAAUUCGCAUCA-AACCGCAGC ....(((((((.(((((..------..))))).))....((((((....)))))).-....))))) ( -19.60, z-score = -2.59, R) >droEre2.scaffold_4820 1248724 60 - 10470090 GAGCGUUGCGUUAGUACUG------UAGUAUUUGCCUCACAUGCGAAUUCGCAUCGCAACCGCAGA ..(((((((((.(((((..------..))))).)).....(((((....))))).)))).)))... ( -18.80, z-score = -1.69, R) >droMoj3.scaffold_6540 1411153 50 - 34148556 ---------GAGAAU-CUG-----UUAUUAUUUGAUCUAUUUGUGUGGCUGCUAUA-GACGCCAGC ---------..((((-..(-----(((.....))))..))))...((((..(....-)..)))).. ( -5.70, z-score = 1.18, R) >consensus GAGCGUUGCGUUAGUACUG______UAGUAUUUGCCUCAGAUGCGAGUUCGCAUCA_AACCGCAGC ....(((((((.(((((((......))))))).))....((((((....))))))......))))) (-12.70 = -14.03 + 1.34)
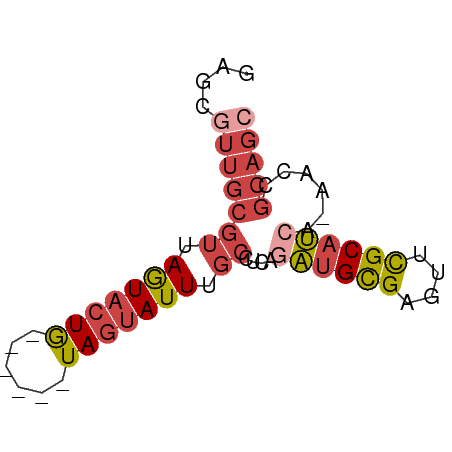
| Location | 18,848,228 – 18,848,287 |
|---|---|
| Length | 59 |
| Sequences | 6 |
| Columns | 66 |
| Reading direction | reverse |
| Mean pairwise identity | 76.19 |
| Shannon entropy | 0.43090 |
| G+C content | 0.48603 |
| Mean single sequence MFE | -14.85 |
| Consensus MFE | -8.88 |
| Energy contribution | -10.77 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.597295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18848228 59 - 27905053 GCUGCGGUU-UGAUGCGAACUCGCAUCUGAGGCAAAUACUA------CAGUACUAACGCAACGCUC (((((((((-(((((((....))))))..))))...(((..------..)))....))))..)).. ( -16.20, z-score = -1.63, R) >droSim1.chr3R 18662300 59 - 27517382 GCUGCGGUU-UGAUGCGAACUCGCAUCUGAGGCAAAUACUA------CAGUACUAACGCAACGCUC (((((((((-(((((((....))))))..))))...(((..------..)))....))))..)).. ( -16.20, z-score = -1.63, R) >droSec1.super_0 19211881 65 - 21120651 GCUGCGGUU-UGAUGCGAACUCGCAUCUGAGGCAAAUACUAACGCACUAGUACUAACGCAACGCUC (((((((((-(((((((....))))))..))))...(((((......)))))....))))..)).. ( -18.60, z-score = -1.97, R) >droYak2.chr3R 19710255 59 - 28832112 GCUGCGGUU-UGAUGCGAAUUCGCAUCUGAGGCAAAUACUA------CAGUACUAACGCAACGCUC (((((((((-(((((((....))))))..))))...(((..------..)))....))))..)).. ( -16.20, z-score = -1.59, R) >droEre2.scaffold_4820 1248724 60 + 10470090 UCUGCGGUUGCGAUGCGAAUUCGCAUGUGAGGCAAAUACUA------CAGUACUAACGCAACGCUC ...(((.((((((((((....)))))((.((.......)))------)........)))))))).. ( -17.30, z-score = -1.23, R) >droMoj3.scaffold_6540 1411153 50 + 34148556 GCUGGCGUC-UAUAGCAGCCACACAAAUAGAUCAAAUAAUAA-----CAG-AUUCUC--------- ..((((...-.......)))).....................-----...-......--------- ( -4.60, z-score = 0.11, R) >consensus GCUGCGGUU_UGAUGCGAACUCGCAUCUGAGGCAAAUACUA______CAGUACUAACGCAACGCUC ..(((((((..((((((....))))))...)))...((((........))))....))))...... ( -8.88 = -10.77 + 1.89)

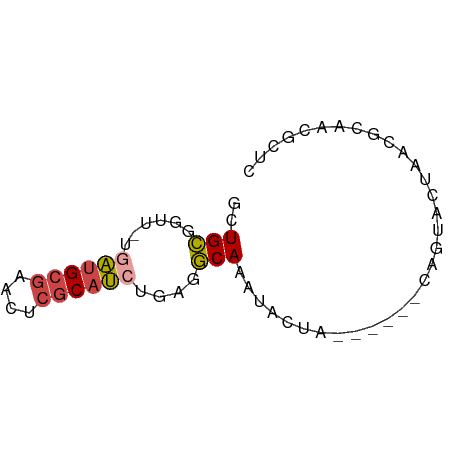
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:37:03 2011