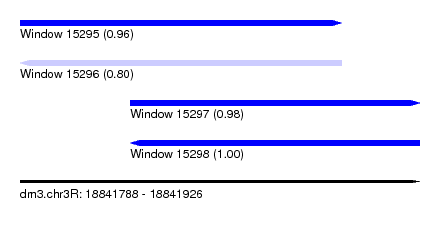
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,841,788 – 18,841,926 |
| Length | 138 |
| Max. P | 0.995690 |

| Location | 18,841,788 – 18,841,899 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 94.26 |
| Shannon entropy | 0.10340 |
| G+C content | 0.55716 |
| Mean single sequence MFE | -45.70 |
| Consensus MFE | -40.92 |
| Energy contribution | -40.80 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.961541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18841788 111 + 27905053 UCCGCGUCCAGUCCAGCUAGUGCAAUGCAGAAAUUGAUUUGCUGCUGCUCCUGCUGAUGCUGCCACUUCUGAUGGCGGUGCAGUAUUGUGGGUGCGGAUUAGUAGGAAUGA (((((..(((...((((..(.(((..(((((......)))))...))).)..))))..((((((((....).))))))).........)))..)))))............. ( -39.80, z-score = -0.96, R) >droSim1.chr3R 18655820 109 + 27517382 UCCGCGUCCAGU--AGCGAGUGCGAUGCAGAAAUUGAUUUGCUGCUGCUCCUGCUGUUGCUGCCACUUCUGAUGGCGGUGCAGCAUUGUGGGUGCGGAUUAGUCGGAAUGG (((((..(((((--(((((((.((((......))))))))))))))((...((((((.((((((((....).)))))))))))))..))))..)))))............. ( -47.40, z-score = -2.71, R) >droSec1.super_0 19205362 109 + 21120651 UCCGCGUCCAGU--AGCGAGUGCGAUGCAGAAAUUGAUUUGCUGCUGCUCCUGCUGUUGCUGCCACUUCUGAUGGCGGUGCAGCAUUGUGGGUGCGGAUUAGUCGGAAUGG (((((..(((((--(((((((.((((......))))))))))))))((...((((((.((((((((....).)))))))))))))..))))..)))))............. ( -47.40, z-score = -2.71, R) >droYak2.chr3R 19703152 109 + 28832112 UCCGCGUCCAGU--GGCGAGUGCGAUGCAGAAAUUGAUUUGCCGCUGCUCCUGCUGUUGCUGCCACUUCUGAUGGCGGUGCAGCAUUGUGGGUGCGGAUUAGUCGGAAUGG (((((..(((((--(((((((.((((......))))))))))))))((...((((((.((((((((....).)))))))))))))..))))..)))))............. ( -49.00, z-score = -2.83, R) >droEre2.scaffold_4820 1242387 106 - 10470090 UCCACGUCCAGU--AGCGAGUGCGAUGCAGAAAUUGAUUUGC---UGCCCCUGCUGUUGCUGCCACUUCUGAUGGCGGUGCAGCAUUGUGGGCGCGGAUUAGUCGGAAUGG (((.((((((((--(((((((.((((......))))))))))---)))...((((((.((((((((....).)))))))))))))...)))))).)))............. ( -44.90, z-score = -2.42, R) >consensus UCCGCGUCCAGU__AGCGAGUGCGAUGCAGAAAUUGAUUUGCUGCUGCUCCUGCUGUUGCUGCCACUUCUGAUGGCGGUGCAGCAUUGUGGGUGCGGAUUAGUCGGAAUGG ((((((((((((..(((((((.((((......)))))))))))...))...(((((.(((((((((....).)))))))))))))...))))))))))............. (-40.92 = -40.80 + -0.12)

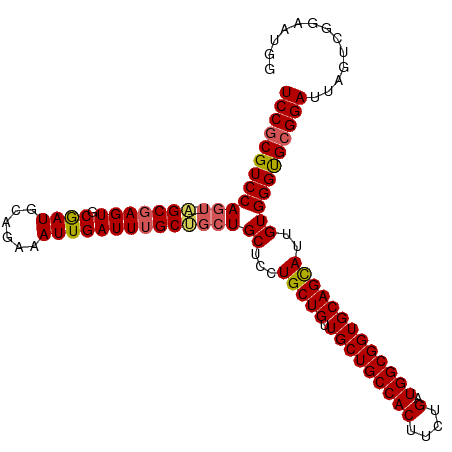
| Location | 18,841,788 – 18,841,899 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 94.26 |
| Shannon entropy | 0.10340 |
| G+C content | 0.55716 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -30.52 |
| Energy contribution | -31.04 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.798933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18841788 111 - 27905053 UCAUUCCUACUAAUCCGCACCCACAAUACUGCACCGCCAUCAGAAGUGGCAGCAUCAGCAGGAGCAGCAGCAAAUCAAUUUCUGCAUUGCACUAGCUGGACUGGACGCGGA .............(((((..(((......(((...(((((.....))))).)))(((((.((.(((((((.(((....)))))))..))).)).)))))..)))..))))) ( -34.10, z-score = -1.81, R) >droSim1.chr3R 18655820 109 - 27517382 CCAUUCCGACUAAUCCGCACCCACAAUGCUGCACCGCCAUCAGAAGUGGCAGCAACAGCAGGAGCAGCAGCAAAUCAAUUUCUGCAUCGCACUCGCU--ACUGGACGCGGA .............(((((..(((...((((((.(((((((.....))))).((....)).)).))))))(((..........)))...((....)).--..)))..))))) ( -34.80, z-score = -2.07, R) >droSec1.super_0 19205362 109 - 21120651 CCAUUCCGACUAAUCCGCACCCACAAUGCUGCACCGCCAUCAGAAGUGGCAGCAACAGCAGGAGCAGCAGCAAAUCAAUUUCUGCAUCGCACUCGCU--ACUGGACGCGGA .............(((((..(((...((((((.(((((((.....))))).((....)).)).))))))(((..........)))...((....)).--..)))..))))) ( -34.80, z-score = -2.07, R) >droYak2.chr3R 19703152 109 - 28832112 CCAUUCCGACUAAUCCGCACCCACAAUGCUGCACCGCCAUCAGAAGUGGCAGCAACAGCAGGAGCAGCGGCAAAUCAAUUUCUGCAUCGCACUCGCC--ACUGGACGCGGA .............(((((..(((....(((((.(((((((.....))))).((....)).)).)))))(((.......................)))--..)))..))))) ( -34.80, z-score = -1.63, R) >droEre2.scaffold_4820 1242387 106 + 10470090 CCAUUCCGACUAAUCCGCGCCCACAAUGCUGCACCGCCAUCAGAAGUGGCAGCAACAGCAGGGGCA---GCAAAUCAAUUUCUGCAUCGCACUCGCU--ACUGGACGUGGA .............((((((.(((...((((((.(((((((.....))))).((....)).)).)))---)))................((....)).--..))).)))))) ( -35.00, z-score = -1.53, R) >consensus CCAUUCCGACUAAUCCGCACCCACAAUGCUGCACCGCCAUCAGAAGUGGCAGCAACAGCAGGAGCAGCAGCAAAUCAAUUUCUGCAUCGCACUCGCU__ACUGGACGCGGA .............(((((..(((...((((((.(((((((.....))))).((....)).)).))))))(((..........)))...((....)).....)))..))))) (-30.52 = -31.04 + 0.52)

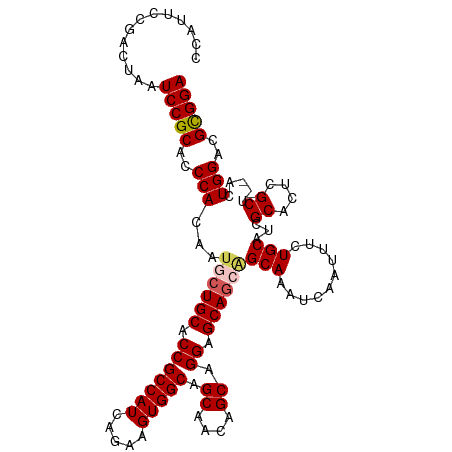
| Location | 18,841,826 – 18,841,926 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 75.00 |
| Shannon entropy | 0.47571 |
| G+C content | 0.56199 |
| Mean single sequence MFE | -38.40 |
| Consensus MFE | -33.29 |
| Energy contribution | -33.55 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.982994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18841826 100 + 27905053 ---UUGCUGCUGCUCC-UGCUGAUGCUGCCACUUCUGAUGGCGGUGCAGUAUUGUGGGUGCGGAUUAGUAGGAAUGACCGGAAUAAACGGCCGUAUCCCGUGGU ---.(((((((((.((-..(.((((((((.(((.(.....).))))))))))))..)).))))..)))))((((((.(((.......))).))).)))...... ( -35.90, z-score = -1.17, R) >droSim1.chr3R 18655856 100 + 27517382 ---UUGCUGCUGCUCC-UGCUGUUGCUGCCACUUCUGAUGGCGGUGCAGCAUUGUGGGUGCGGAUUAGUCGGAAUGGCCGGAAUAAGCGGCCGUAUCCCGUGGU ---(..(..((((.((-..(...((((((.(((.(.....).)))))))))..)..)).)))).......((((((((((.......))))))).))).)..). ( -40.90, z-score = -1.09, R) >droSec1.super_0 19205398 100 + 21120651 ---UUGCUGCUGCUCC-UGCUGUUGCUGCCACUUCUGAUGGCGGUGCAGCAUUGUGGGUGCGGAUUAGUCGGAAUGGCCGGAAUAAGCGGCCGUAUCCCGUGGU ---(..(..((((.((-..(...((((((.(((.(.....).)))))))))..)..)).)))).......((((((((((.......))))))).))).)..). ( -40.90, z-score = -1.09, R) >droYak2.chr3R 19703188 100 + 28832112 ---UUGCCGCUGCUCC-UGCUGUUGCUGCCACUUCUGAUGGCGGUGCAGCAUUGUGGGUGCGGAUUAGUCGGAAUGGCCGGAAUAAACGGCCGUAUCCCGUGGU ---..((((((((.((-..(...((((((.(((.(.....).)))))))))..)..)).)))........((((((((((.......))))))).))).))))) ( -44.30, z-score = -2.30, R) >droEre2.scaffold_4820 1242423 97 - 10470090 ------UUGCUGCCCC-UGCUGUUGCUGCCACUUCUGAUGGCGGUGCAGCAUUGUGGGCGCGGAUUAGUCGGAAUGGCCGGAAUAAACGGCCAUAUCCCGUGGU ------((((.(((((-((((((.((((((((....).)))))))))))))..).)))))))).......((((((((((.......))))))).)))...... ( -44.10, z-score = -2.44, R) >droVir3.scaffold_12822 4027219 84 - 4096053 UUUCUGAUGAUGCUUUGUGAUGCUGCUGCUGCCGUCGUCAACAGUGCAGUAGC-CAGGCUUCGACAAACUUUAAUUACAGACAAC------------------- ..((((.((((..(((((((.((((((((((((..........).))))))))-..))).)).))))).....))))))))....------------------- ( -24.30, z-score = -1.29, R) >consensus ___UUGCUGCUGCUCC_UGCUGUUGCUGCCACUUCUGAUGGCGGUGCAGCAUUGUGGGUGCGGAUUAGUCGGAAUGGCCGGAAUAAACGGCCGUAUCCCGUGGU ......((((.((((..((((((.((((((((....).)))))))))))))....)))))))).......((((((((((.......))))))).)))...... (-33.29 = -33.55 + 0.26)

| Location | 18,841,826 – 18,841,926 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 75.00 |
| Shannon entropy | 0.47571 |
| G+C content | 0.56199 |
| Mean single sequence MFE | -32.95 |
| Consensus MFE | -24.78 |
| Energy contribution | -26.57 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.995690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18841826 100 - 27905053 ACCACGGGAUACGGCCGUUUAUUCCGGUCAUUCCUACUAAUCCGCACCCACAAUACUGCACCGCCAUCAGAAGUGGCAGCAUCAGCA-GGAGCAGCAGCAA--- .....((((...(((((.......)))))..))))....................((((.(((((((.....))))).((....)).-)).))))......--- ( -27.50, z-score = -1.41, R) >droSim1.chr3R 18655856 100 - 27517382 ACCACGGGAUACGGCCGCUUAUUCCGGCCAUUCCGACUAAUCCGCACCCACAAUGCUGCACCGCCAUCAGAAGUGGCAGCAACAGCA-GGAGCAGCAGCAA--- ....((((((..(((((.......)))))))))))..................((((((.(((((((.....))))).((....)).-)).))))))....--- ( -37.00, z-score = -3.30, R) >droSec1.super_0 19205398 100 - 21120651 ACCACGGGAUACGGCCGCUUAUUCCGGCCAUUCCGACUAAUCCGCACCCACAAUGCUGCACCGCCAUCAGAAGUGGCAGCAACAGCA-GGAGCAGCAGCAA--- ....((((((..(((((.......)))))))))))..................((((((.(((((((.....))))).((....)).-)).))))))....--- ( -37.00, z-score = -3.30, R) >droYak2.chr3R 19703188 100 - 28832112 ACCACGGGAUACGGCCGUUUAUUCCGGCCAUUCCGACUAAUCCGCACCCACAAUGCUGCACCGCCAUCAGAAGUGGCAGCAACAGCA-GGAGCAGCGGCAA--- .((.((((((..(((((.......)))))))))))...................(((((.(((((((.....))))).((....)).-)).)))))))...--- ( -37.90, z-score = -3.14, R) >droEre2.scaffold_4820 1242423 97 + 10470090 ACCACGGGAUAUGGCCGUUUAUUCCGGCCAUUCCGACUAAUCCGCGCCCACAAUGCUGCACCGCCAUCAGAAGUGGCAGCAACAGCA-GGGGCAGCAA------ ....(((((..((((((.......)))))))))))........((((((....(((((..(.(((((.....))))).)...)))))-.)))).))..------ ( -39.90, z-score = -3.12, R) >droVir3.scaffold_12822 4027219 84 + 4096053 -------------------GUUGUCUGUAAUUAAAGUUUGUCGAAGCCUG-GCUACUGCACUGUUGACGACGGCAGCAGCAGCAUCACAAAGCAUCAUCAGAAA -------------------....((((.........(((((.((.((...-(((.((((..((....))...)))).))).)).))))))).......)))).. ( -18.39, z-score = 1.05, R) >consensus ACCACGGGAUACGGCCGUUUAUUCCGGCCAUUCCGACUAAUCCGCACCCACAAUGCUGCACCGCCAUCAGAAGUGGCAGCAACAGCA_GGAGCAGCAGCAA___ ....((((((..(((((.......)))))))))))..................((((((.(((((((.....))))).((....))..)).))))))....... (-24.78 = -26.57 + 1.78)
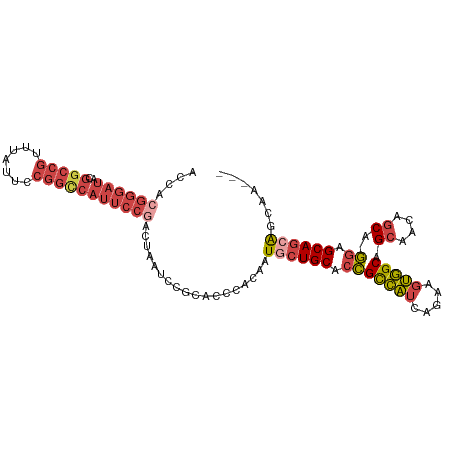
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:37:01 2011