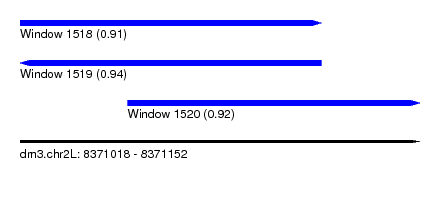
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,371,018 – 8,371,152 |
| Length | 134 |
| Max. P | 0.943596 |

| Location | 8,371,018 – 8,371,119 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 57.17 |
| Shannon entropy | 0.78072 |
| G+C content | 0.55924 |
| Mean single sequence MFE | -18.05 |
| Consensus MFE | -9.25 |
| Energy contribution | -8.03 |
| Covariance contribution | -1.22 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.61 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905739 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
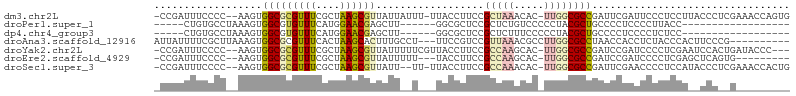
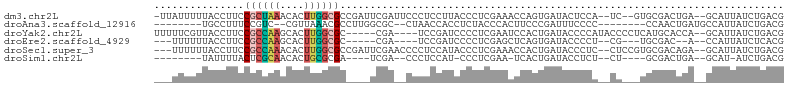
>dm3.chr2L 8371018 101 + 23011544 -CCGAUUUCCCC--AAGUGGCGCGUUUCGCUAAGCGUUAUUAUUU-UUACCUUCCGCUAAACAC-UUGGCGCCGAUUCGAUUCCCUCCUUACCCUCGAAACCAGUG -.........((--((((((((.....)))..((((.........-........))))...)))-))))(((.(.(((((..............))))).)..))) ( -17.17, z-score = -0.28, R) >droPer1.super_1 2172231 77 + 10282868 -----CUGUGCCUAAAGUGGCGUGUUUCAUGGAACGAGCUU------GGCGCUCCGCUCUGUCCCCCUACGCUGCCCCUCCCCUUACC------------------ -----..(((.....((.(((((((.....(((..((((..------((....))))))..)))....)))).))).)).....))).------------------ ( -17.40, z-score = -0.23, R) >dp4.chr4_group3 696081 77 + 11692001 -----CUGUGCCUAAAGUGGCGUGUUUCAUGGAACGAGCUU------GGCGCUCCGCUCUUUCCCCCUACGCUGCCCCUCCCCUCUCC------------------ -----..........((.(((((((.....((((.((((..------((....)))))).))))....)))).))).)).........------------------ ( -18.80, z-score = -1.15, R) >droAna3.scaffold_12916 5455602 93 + 16180835 AUUAUUUUCGCUUAAAGUGGCGCGUUUCACUAAGCACUUUGCCU---UUCCGUCCGUUAAACGCCUUGGCGCCUAACCACCUCUACCCACUUCCCG---------- ..............((((((.((..........)).........---........((((..(((....)))..)))).........))))))....---------- ( -14.60, z-score = -0.42, R) >droYak2.chr2L 11009140 99 + 22324452 -CCGAUUUCCCC--AAGUGGCGCGUUUCGCUAAGCGUUAUUUUUCGUUACCUUCCGCCAAGCAC-UUGGCGCCGAUCCGAUCCCCUCGAAUCCACUGAUACCC--- -...........--.((((((((((((....))))))..................(((((....-))))))))(((.(((.....))).))).))).......--- ( -20.00, z-score = -0.53, R) >droEre2.scaffold_4929 17280496 90 + 26641161 -CCGAUUUCCCC--AAGUGGCGCGUUUCGCUAAGCGUUAUUUUU---UACCUUCCGCCAAGCAC-UUGGCGCCGAUCCGAUCCCCUCGAGCUCAGUG--------- -..((.......--(((((((((..........)))))))))..---.....))((((((....-))))))..((.((((.....))).).))....--------- ( -19.04, z-score = -0.01, R) >droSec1.super_3 3867605 99 + 7220098 -CCGAUUUCCCC--AAGUGGCGCGUUUCGCUAAGCGUUAUU--UU-UUACCUUCCGCCAAACAC-UUGGCGCCGAUUCGAACCCCUCCAUACCCUCGAAACCACUG -...........--.(((((.((((((....))))))....--..-........((((((....-))))))....(((((..............))))).))))). ( -19.34, z-score = -1.66, R) >consensus _CCGAUUUCCCC__AAGUGGCGCGUUUCGCUAAGCGUUAUU__U___UACCUUCCGCUAAACAC_UUGGCGCCGAUCCGACCCCCUCCA__CCC__G_________ ..................(((((((((....))))....................(.....)......)))))................................. ( -9.25 = -8.03 + -1.22)
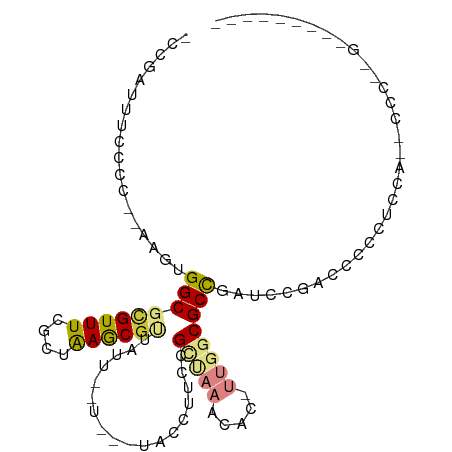
| Location | 8,371,018 – 8,371,119 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 57.17 |
| Shannon entropy | 0.78072 |
| G+C content | 0.55924 |
| Mean single sequence MFE | -26.78 |
| Consensus MFE | -12.36 |
| Energy contribution | -12.06 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.943596 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 8371018 101 - 23011544 CACUGGUUUCGAGGGUAAGGAGGGAAUCGAAUCGGCGCCAA-GUGUUUAGCGGAAGGUAA-AAAUAAUAACGCUUAGCGAAACGCGCCACUU--GGGGAAAUCGG- ..((((((((..((((...((.....)).....(((((...-.((((.((((........-.........)))).))))....)))))))))--...))))))))- ( -28.03, z-score = -1.16, R) >droPer1.super_1 2172231 77 - 10282868 ------------------GGUAAGGGGAGGGGCAGCGUAGGGGGACAGAGCGGAGCGCC------AAGCUCGUUCCAUGAAACACGCCACUUUAGGCACAG----- ------------------......((((.((((.((....(....)...))((....))------..)))).)))).........(((......)))....----- ( -23.50, z-score = -0.98, R) >dp4.chr4_group3 696081 77 - 11692001 ------------------GGAGAGGGGAGGGGCAGCGUAGGGGGAAAGAGCGGAGCGCC------AAGCUCGUUCCAUGAAACACGCCACUUUAGGCACAG----- ------------------...(...((((.(((.(.((....((((.((((((....))------..)))).)))).....)).)))).))))...)....----- ( -19.90, z-score = 0.33, R) >droAna3.scaffold_12916 5455602 93 - 16180835 ----------CGGGAAGUGGGUAGAGGUGGUUAGGCGCCAAGGCGUUUAACGGACGGAA---AGGCAAAGUGCUUAGUGAAACGCGCCACUUUAAGCGAAAAUAAU ----------((.(((((((((...(.(.(((((((((....))))))))).).)....---((((.....))))........)).)))))))...))........ ( -26.70, z-score = -1.60, R) >droYak2.chr2L 11009140 99 - 22324452 ---GGGUAUCAGUGGAUUCGAGGGGAUCGGAUCGGCGCCAA-GUGCUUGGCGGAAGGUAACGAAAAAUAACGCUUAGCGAAACGCGCCACUU--GGGGAAAUCGG- ---.(((.((....(((((((.....)))))))(((((...-.((((.((((..................)))).))))....)))))....--...)).)))..- ( -31.67, z-score = -1.45, R) >droEre2.scaffold_4929 17280496 90 - 26641161 ---------CACUGAGCUCGAGGGGAUCGGAUCGGCGCCAA-GUGCUUGGCGGAAGGUA---AAAAAUAACGCUUAGCGAAACGCGCCACUU--GGGGAAAUCGG- ---------..((((.(((.(((.(((...)))(((((...-.((((.((((.......---........)))).))))....))))).)))--.)))...))))- ( -29.86, z-score = -1.45, R) >droSec1.super_3 3867605 99 - 7220098 CAGUGGUUUCGAGGGUAUGGAGGGGUUCGAAUCGGCGCCAA-GUGUUUGGCGGAAGGUAA-AA--AAUAACGCUUAGCGAAACGCGCCACUU--GGGGAAAUCGG- (.((..(..(((((((.....((.(((......))).))..-((((((.((..(((((..-..--....)).))).)).))))))))).)))--)..)..)).).- ( -27.80, z-score = -0.65, R) >consensus _________C__GGG__UGGAGGGGAUCGGAUCGGCGCCAA_GUGCUUAGCGGAAGGUA___A__AAUAACGCUUAGCGAAACGCGCCACUU__GGGGAAAUCGG_ .................................(((((.....((((.((((..................)))).))))....))))).................. (-12.36 = -12.06 + -0.30)



| Location | 8,371,054 – 8,371,152 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 59.93 |
| Shannon entropy | 0.72874 |
| G+C content | 0.52962 |
| Mean single sequence MFE | -13.90 |
| Consensus MFE | -4.81 |
| Energy contribution | -4.87 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.916850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8371054 98 + 23011544 -UUAUUUUUACCUUCCGCUAAACACUUGGCGCCGAUUCGAUUCCCUCCUUACCCUCGAAACCAGUGAUACUCCA--UC--GUGCGACUGA--GCAUUAUCUGACG -..............((((((....))))))....(((((..............)))))..(((((((.(((..--((--....))..))--).)))).)))... ( -15.34, z-score = -1.15, R) >droAna3.scaffold_12916 5455641 85 + 16180835 --------UGCCUUUCCGUC--CGUUAAACGCCUUGGCGC--CUAACCACCUCUACCCACUUCCCGAUUUCCCC--------CCAACUGAUGCCAUUAUCUGACG --------........((((--.((((..(((....))).--.))))...........................--------......((((....)))).)))) ( -10.70, z-score = -1.93, R) >droYak2.chr2L 11009176 94 + 22324452 UUUUUCGUUACCUUCCGCCAAGCACUUGGCGC-----CGA----UCCGAUCCCCUCGAAUCCACUGAUACCCCAUACCCCUCAUGCACCA--GCAUUAUCUGACG .....(((((.....((((((....)))))).-----.((----(.(((.....))).))).....................((((....--))))....))))) ( -15.90, z-score = -2.40, R) >droEre2.scaffold_4929 17280532 84 + 26641161 ---UUUUUUACCUUCCGCCAAGCACUUGGCGC-----CGA----UCCGAUCCCCUCGAGCUCAGUGAUACCCCU--CG---UGCGAC--A--CCAUUAUCUCACG ---....((((....((((((....)))))).-----.((----.((((.....))).).)).)))).......--((---((.((.--.--......)).)))) ( -15.60, z-score = -1.43, R) >droSec1.super_3 3867641 98 + 7220098 ---UUUUUUACCUUCCGCCAAACACUUGGCGCCGAUUCGAACCCCUCCAUACCCUCGAAACCACUGAUACCCUC--CUCCGUGCGACAGA--GCAUUAUCUGACG ---............((((((....)))))).((.(((((..............)))))..(((.((.......--.)).))))).((((--......))))... ( -16.14, z-score = -2.64, R) >droSim1.chr2L 8156987 80 + 22036055 --------UAUUUUACUCGCAACACUGCGCGA----UCGA--CCCUCCAU-CCCUCGAA-UCACUGAUACCUCU--CU----GCGACUGA--GCAU-AUCUGACG --------.......(.((((....)))).).----((((--........-...)))).-(((..((((...((--(.----......))--)..)-)))))).. ( -9.70, z-score = -0.38, R) >consensus ___UUUUUUACCUUCCGCCAAACACUUGGCGC____UCGA__CCCUCCAUCCCCUCGAAACCACUGAUACCCCA__CC___UGCGACUGA__GCAUUAUCUGACG ...............((((((....)))))).......................................................................... ( -4.81 = -4.87 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:25:50 2011