
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,830,511 – 18,830,580 |
| Length | 69 |
| Max. P | 0.719176 |

| Location | 18,830,511 – 18,830,580 |
|---|---|
| Length | 69 |
| Sequences | 11 |
| Columns | 83 |
| Reading direction | reverse |
| Mean pairwise identity | 70.65 |
| Shannon entropy | 0.61436 |
| G+C content | 0.38306 |
| Mean single sequence MFE | -20.42 |
| Consensus MFE | -5.83 |
| Energy contribution | -5.84 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.719176 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
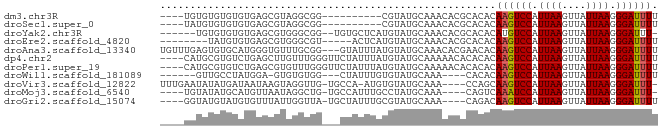
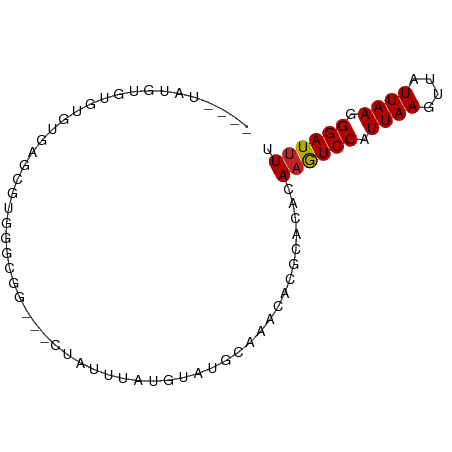
>dm3.chr3R 18830511 69 - 27905053 ----UGUGUGUGUGUGAGCGUAGGCGG----------CGUAUGCAAACACGCACACAAGUCCAUUAAGUUAUUAAGGGAUUUU ----.((((((((((..(((((.(...----------).)))))..))))))))))((((((.((((....)))).)))))). ( -28.50, z-score = -4.69, R) >droSec1.super_0 19194286 69 - 21120651 ----UAUGUGUGUGUGAGCGUAGGCGG----------CGUAUGCAAACACGCACACAAGUCCAUUAAGUUAUUAAGGGAUUUU ----..(((((((((..(((((.(...----------).)))))..))))))))).((((((.((((....)))).)))))). ( -24.90, z-score = -3.74, R) >droYak2.chr3R 19691555 74 - 28832112 ------UGUGUGUGUGAGCGUGGGCGG--UGUGCUCAUGUAUGCAAACACGCACACAUGUCCAUUAAGUUAUUAAGGGAUUU- ------((((((((((.((((((((..--...)))))))).......)))))))))).((((.((((....)))).))))..- ( -27.30, z-score = -2.50, R) >droEre2.scaffold_4820 1231290 70 + 10470090 --------UAUGUGUGAGCGUGGGCGU-----ACUCAUGUAUGCAAACACGCACACAAGUCCAUUAAGUUAUUAAGGGAUUUU --------....((((.(((((.((((-----((....))))))...))))).))))(((((.((((....)))).))))).. ( -27.30, z-score = -4.91, R) >droAna3.scaffold_13340 11208268 80 + 23697760 UGUUUGAGUGUGCAUGGGUGUUUGCGG---GUAUUUAUGUAUGCAAACACGAACACAAGUCCAUUAAGUUAUUAAGGGAUUUU ((((((.((.((((((.(((..((...---.))..))).)))))).)).)))))).((((((.((((....)))).)))))). ( -22.80, z-score = -2.69, R) >dp4.chr2 24747158 79 + 30794189 ----CAUGCGUGUCUGAGCUUGUUUGGGUUCUAUUUAUGUAUGCAAAAACACACACAAGUCCAUUAAGUUAUUAAGGGAUUUU ----((((((((...((((((....))))))....)))))))).............((((((.((((....)))).)))))). ( -18.10, z-score = -1.46, R) >droPer1.super_19 84288 79 + 1869541 ----CAUGCGUGUCUGAGCGUGUUUGGGUUCUAUUUAUGUAUGCAAAAACACACACAAGUCCAUUAAGUUAUUAAGGGAUUUU ----((((((((...((((.(....).))))....)))))))).............((((((.((((....)))).)))))). ( -16.50, z-score = -0.69, R) >droWil1.scaffold_181089 11167621 69 + 12369635 ------GUUGCCUAUGGA-GUGUGUGG---CUAUUUGUGUAUGCAAA----CACACAAGUCCAUUAAGUUAUUAAGGGAUUUU ------....(((.((((-.((((((.---...(((((....)))))----))))))..))))((((....)))))))..... ( -15.80, z-score = -0.60, R) >droVir3.scaffold_12822 4017032 76 + 4096053 UUUGAAUAUAUGAUAAUAAGUAGGUUG-UGCCA-AUGUGUAUGCAAA----CCAGCAAGUCCAUUAAGUUAUUAAGGGAUUU- ...................((.((((.-(((..-........)))))----)).))((((((.((((....)))).))))))- ( -12.50, z-score = -0.19, R) >droMoj3.scaffold_6540 1388782 73 + 34148556 ----UGUAUAUGCAUGUUAAUAGGCUG-UGCCAUUUGCCUAUGCAAA----CAGUCAAAUCCAUUAAGUUAUUAAGGGAUUU- ----..........((((.((((((..-........))))))...))----))...((((((.((((....)))).))))))- ( -15.40, z-score = -0.47, R) >droGri2.scaffold_15074 3639235 74 - 7742996 ----GGUAUGUAUGUGUUUAUUGGUUA-UGCUAUUUGCGUAUGCAAA----CAGACAAGUCCAUUAAGUUAUUAAGGGAUUUU ----.((.(((((((((....(((...-..)))...))))))))).)----)....((((((.((((....)))).)))))). ( -15.50, z-score = -1.11, R) >consensus ____UAUGUGUGUGUGAGCGUGGGCGG___CUAUUUAUGUAUGCAAACACGCACACAAGUCCAUUAAGUUAUUAAGGGAUUUU ........................................................((((((.((((....)))).)))))). ( -5.83 = -5.84 + 0.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:36:58 2011