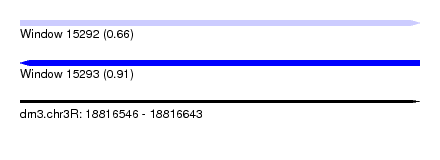
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,816,546 – 18,816,643 |
| Length | 97 |
| Max. P | 0.907879 |

| Location | 18,816,546 – 18,816,643 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 59.40 |
| Shannon entropy | 0.79897 |
| G+C content | 0.59461 |
| Mean single sequence MFE | -25.29 |
| Consensus MFE | -7.89 |
| Energy contribution | -8.87 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.661405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
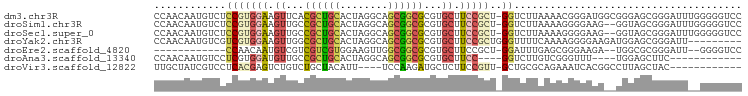
>dm3.chr3R 18816546 97 + 27905053 GGACCCCCAAAUCCCGCUCCCGCCAUCCCGUUUUAAGACC-AGCGGAAGCACGCGCCGCUGCCUAGUGCAGCGUGAACUUCCACGGAGACAUUGUUGG ......((((......((((.........(((....))).-.(.(((((..(((((.((........)).)))))..))))).)))))......)))) ( -28.20, z-score = -0.92, R) >droSim1.chr3R 18630647 95 + 27517382 GGACCCCCAAAUCCCGCUACC--CUUCCCCUUUUAAGACC-AGCGGAAGCACGCGCCGCUGCCUAGUGCAGCGGCAACUUCCACGGAGACAUUGUUGG (((........))).......--...............((-((((((((.....((((((((.....))))))))..)))))..(....)...))))) ( -31.20, z-score = -2.35, R) >droSec1.super_0 19180510 95 + 21120651 GGACCCCCAAAUCCCGCUACC--CUUCCCCUUUUAAGACC-AGCGGAAGCACGCGCCGCUGCCUAGUGCAGCGGCAACUUCCACGGAGACAUUGUUGG (((........))).......--...............((-((((((((.....((((((((.....))))))))..)))))..(....)...))))) ( -31.20, z-score = -2.35, R) >droYak2.chr3R 19676611 89 + 28832112 ---------AAUCCCGCUCCAUCUUCCCCUUUUGAAAACCCAGCGGAAGCACGCGCCGCUGCCUAGUGCAGCGCCAACUUCCACGACGACAUUGUUGG ---------......(((.....(((.......))).....)))(((((...((((.((........)).))))...))))).(((((....))))). ( -21.90, z-score = -1.03, R) >droEre2.scaffold_4820 1217555 81 - 10470090 GGACCCC--AAUCCCGCGCCA--UCUUCCCGCUCAAAUCC-AGCGGAAGCACGCGCCGCCAACUUCCACGACGACGACAUUGUUGG------------ (((....--..))).((((..--.(((((.(((.......-))))))))...))))..(((((.....((....)).....)))))------------ ( -21.30, z-score = -1.46, R) >droAna3.scaffold_13340 11193770 78 - 23697760 ------------GAAGCUCCA----AAACCCGACAAGACC----GGAAGCACGCGCCGCUGCCUAGUGCAGCGGCAACAUCCACGAGGACAUUGUUGG ------------.........----....(((((((..((----(((.(....)((((((((.....))))))))....)))....))...))))))) ( -27.00, z-score = -2.31, R) >droVir3.scaffold_12822 4002777 81 - 4096053 ------------GUAGCUAAGGCCGUGAUUUCUGCGCAGC-AACGGAAGAGCAUCUUGGA----AAUGUAGCAGACAGACUCGUGAGGACGAUAGCAA ------------...((((...........(((((.....-..(....).((((......----.)))).))))).....((((....)))))))).. ( -16.20, z-score = 1.60, R) >consensus GGACCCC__AAUCCCGCUCCA__CUUCCCCUUUUAAGACC_AGCGGAAGCACGCGCCGCUGCCUAGUGCAGCGGCAACUUCCACGGAGACAUUGUUGG ..........................................((....))......((((((.....))))))......................... ( -7.89 = -8.87 + 0.98)

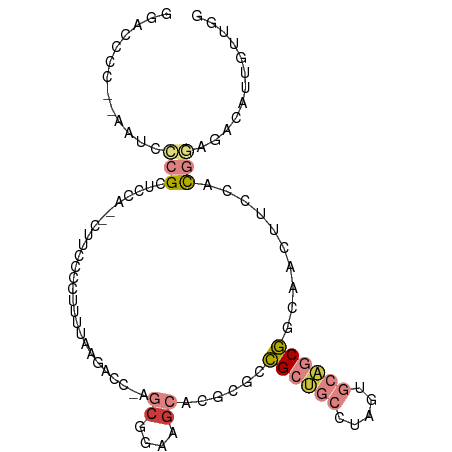
| Location | 18,816,546 – 18,816,643 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 59.40 |
| Shannon entropy | 0.79897 |
| G+C content | 0.59461 |
| Mean single sequence MFE | -33.89 |
| Consensus MFE | -11.69 |
| Energy contribution | -12.49 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.907879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18816546 97 - 27905053 CCAACAAUGUCUCCGUGGAAGUUCACGCUGCACUAGGCAGCGGCGCGUGCUUCCGCU-GGUCUUAAAACGGGAUGGCGGGAGCGGGAUUUGGGGGUCC ((..(((.((((((((((.....).((((((.....)))))).)))).(((((((((-(.((((.....))))))))))))))))))))))..))... ( -40.70, z-score = -1.77, R) >droSim1.chr3R 18630647 95 - 27517382 CCAACAAUGUCUCCGUGGAAGUUGCCGCUGCACUAGGCAGCGGCGCGUGCUUCCGCU-GGUCUUAAAAGGGGAAG--GGUAGCGGGAUUUGGGGGUCC ((..(((.(((.((..(((((((((((((((.....)))))))))...))))))(((-(.((((........)))--).))))))))))))..))... ( -43.70, z-score = -2.83, R) >droSec1.super_0 19180510 95 - 21120651 CCAACAAUGUCUCCGUGGAAGUUGCCGCUGCACUAGGCAGCGGCGCGUGCUUCCGCU-GGUCUUAAAAGGGGAAG--GGUAGCGGGAUUUGGGGGUCC ((..(((.(((.((..(((((((((((((((.....)))))))))...))))))(((-(.((((........)))--).))))))))))))..))... ( -43.70, z-score = -2.83, R) >droYak2.chr3R 19676611 89 - 28832112 CCAACAAUGUCGUCGUGGAAGUUGGCGCUGCACUAGGCAGCGGCGCGUGCUUCCGCUGGGUUUUCAAAAGGGGAAGAUGGAGCGGGAUU--------- ((..(....(((((((((((((..(((((((........)))))))..))))))))....((..(....)..)).))))).)..))...--------- ( -34.10, z-score = -1.57, R) >droEre2.scaffold_4820 1217555 81 + 10470090 ------------CCAACAAUGUCGUCGUCGUGGAAGUUGGCGGCGCGUGCUUCCGCU-GGAUUUGAGCGGGAAGA--UGGCGCGGGAUU--GGGGUCC ------------.(..((((.(((((((((.......)))))))((((.((((((((-.......))).))))).--..)))))).)))--)..)... ( -30.50, z-score = -0.69, R) >droAna3.scaffold_13340 11193770 78 + 23697760 CCAACAAUGUCCUCGUGGAUGUUGCCGCUGCACUAGGCAGCGGCGCGUGCUUCC----GGUCUUGUCGGGUUU----UGGAGCUUC------------ ((((.(((((((....)))))))((((((((.....)))))))).......(((----((.....)))))..)----)))......------------ ( -30.00, z-score = -1.31, R) >droVir3.scaffold_12822 4002777 81 + 4096053 UUGCUAUCGUCCUCACGAGUCUGUCUGCUACAUU----UCCAAGAUGCUCUUCCGUU-GCUGCGCAGAAAUCACGGCCUUAGCUAC------------ ..((((((((....))))((((((((((..((((----.....)))).......((.-...)))))))...)).)))..))))...------------ ( -14.50, z-score = 0.73, R) >consensus CCAACAAUGUCUCCGUGGAAGUUGCCGCUGCACUAGGCAGCGGCGCGUGCUUCCGCU_GGUCUUAAAAGGGGAAG__GGGAGCGGGAUU__GGGGUCC ..............(((((.((...((((((........))))))...)).))))).......................................... (-11.69 = -12.49 + 0.80)
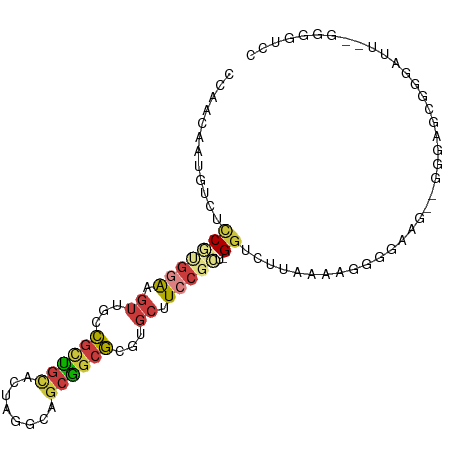
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:36:57 2011