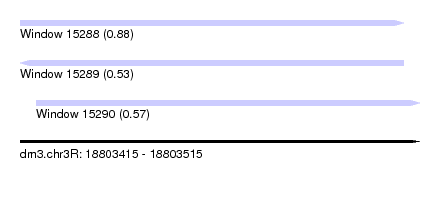
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,803,415 – 18,803,515 |
| Length | 100 |
| Max. P | 0.877159 |

| Location | 18,803,415 – 18,803,511 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 80.40 |
| Shannon entropy | 0.39984 |
| G+C content | 0.57086 |
| Mean single sequence MFE | -36.34 |
| Consensus MFE | -20.65 |
| Energy contribution | -20.34 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.877159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
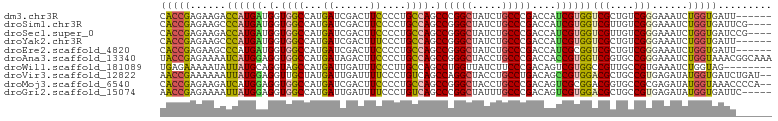
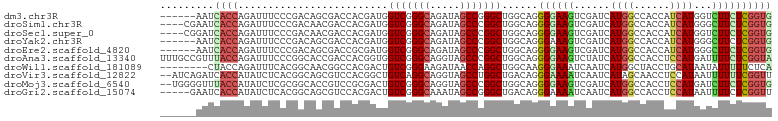
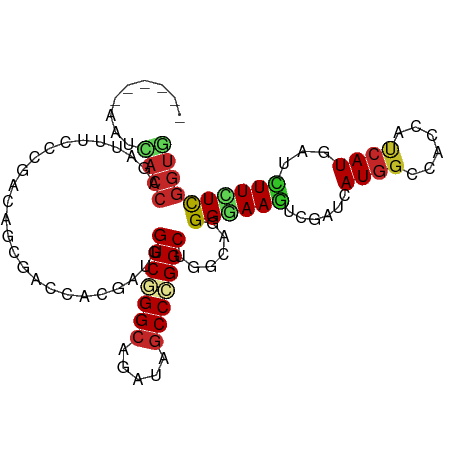
>dm3.chr3R 18803415 96 + 27905053 CACCGAGAAGACCAUGAUGGUGGCCAUGAUCGACUUCCCCUGCCAGCCCGGCUAUCUGCCCGACCAUCGUGGUCGCUGUCGGGAAAUCUGGUGAUU------ ..((((.(.(((((((((((((((...((......))....))).....(((.....)))..))))))))))))..).))))..((((....))))------ ( -37.40, z-score = -2.09, R) >droSim1.chr3R 18616689 98 + 27517382 CACCGAGAAGCCCAUGAUGGUGGCCAUGAUCGACUUCCCCUGCCAGCCGGGCUAUCUGCCCGACCAUCGUGGUCGUUGUCGGGAAAUCUGGUGAUUCG---- ..((((.(((.(((((((((((((...((......))....)))...(((((.....))))))))))))))).).)).))))(((.((....))))).---- ( -39.50, z-score = -2.13, R) >droSec1.super_0 19167546 98 + 21120651 CACCGAGAAGACCAUGAUGGUGGCCAUGAUCGACUUCCCCUGCCAGCCGGGCUAUCUGCCCGACCAUCGUGGUCGUUGUCGGGAAAUCUGGUGAUCCG---- ..((((.(((((((((((((((((...((......))....)))...(((((.....))))))))))))))))).)).)))).......((....)).---- ( -44.30, z-score = -3.50, R) >droYak2.chr3R 19663077 96 + 28832112 CACCGAGAAGCCCAUGAUGGUGGCCAUGAUCGACUUUCCCUGCCAGCCGGGCUAUCUGCCCGACCAUCGUGGUCGCUGUCGGGAAAUCUGGUGAUU------ ((((.(((..(((..(((((((((((((((.(.........(....)(((((.....))))).).))))))))))))))))))...)))))))...------ ( -42.30, z-score = -3.08, R) >droEre2.scaffold_4820 1203635 96 - 10470090 CACCGAGAAGCCCAUGAUGGUGGCCAUGAUCGACUUCCCCUGCCAGCCGGGCUAUCUGCCCGACCAUCGCGGUCGCUGUCGGGAAAUCUGGUGAUU------ (((((.((((.(.((.((((...)))).)).).)))).((((.((((.((((.....))))((((.....)))))))).)))).....)))))...------ ( -39.20, z-score = -1.92, R) >droAna3.scaffold_13340 11179480 102 - 23697760 UACCGAGAAAAUCAUGGAGGUGGCCAUGAUAGACUUCCCCUGCCAGCCGGGCUACCUGCCCGACCACCGUGGUCGGUGCCGGGAAAUCUGGUAAACGGCAAA ((((......(((((((......)))))))(((.(((((..((..(((((((.....))))((((.....))))))))).))))).)))))))......... ( -41.40, z-score = -2.13, R) >droWil1.scaffold_181089 11140380 94 - 12369635 UGAGAAAAAUAUUAUGCAGGUAGCCAUGAUUGAUUUCCCUUGCCAGCCUGGUUAUCUUCCCGACAGUCGUGGCCGUUGCCGUGAAAUCUGGUAG-------- ..................((((((((((((((.........(((.....)))...........))))))))))...))))..............-------- ( -20.85, z-score = 0.01, R) >droVir3.scaffold_12822 3991432 100 - 4096053 AACCGAAAAAAUUAUGGAGGUUGCUAUGAUUGAUUUUCCCUGUCAGCCAGGCUACCUGCCUGACAGCCGUGGACGCUGCCGUGAGAUAUGGUGAUCUGAU-- .((((......((((((.(((..(((((...(.....).(((((((.((((...)))).))))))).)))))..))).))))))....))))........-- ( -28.90, z-score = -1.71, R) >droMoj3.scaffold_6540 1343659 100 - 34148556 CACCGAGAAGAUCAUGGAGGUGGCCAUGAUCGACUUCCCCUGCCAGCCGGGCUACCUGCCCGACAGUCGCGGACGGUGCCGCGAGAUAUGGUAAACCCCA-- ....(((..((((((((......))))))))..)))....((((((.(((((.....))))).)..((((((......))))))....))))).......-- ( -39.80, z-score = -2.06, R) >droGri2.scaffold_15074 3608096 97 + 7742996 AACCGAGAAAAUUAUGGAGGUGGCCAUGAUUGAUUUUCCCUGUCAGCCCGGCUAUUUGCCCGACAGUCGUGGACGCUGCCGUGAGAUAUGGUGAUUC----- .((((......((((((.((((.(((((((((.........(....)..(((.....)))...))))))))).)))).))))))....)))).....----- ( -29.80, z-score = -1.38, R) >consensus CACCGAGAAGACCAUGAAGGUGGCCAUGAUCGACUUCCCCUGCCAGCCGGGCUAUCUGCCCGACCAUCGUGGUCGCUGCCGGGAAAUCUGGUGAUU______ (((((......((((((.(.((((...((......))....)))).)(((((.....)))))....))))))(((....)))......)))))......... (-20.65 = -20.34 + -0.31)
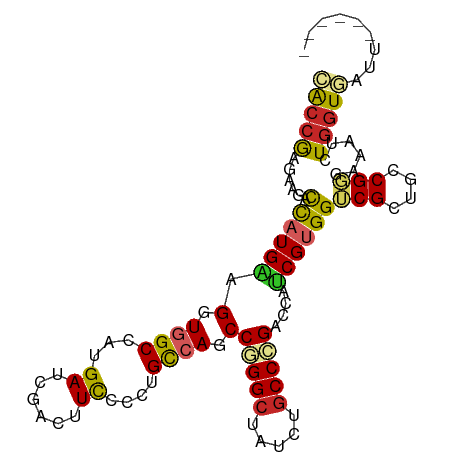
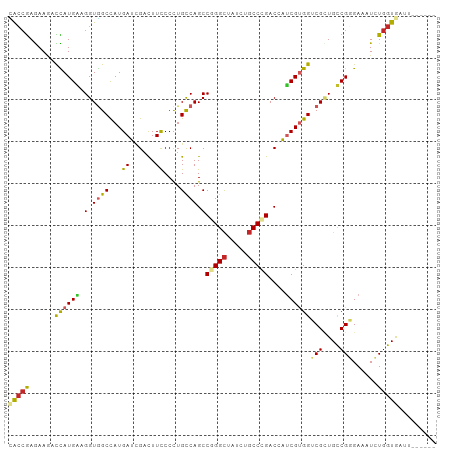
| Location | 18,803,415 – 18,803,511 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 80.40 |
| Shannon entropy | 0.39984 |
| G+C content | 0.57086 |
| Mean single sequence MFE | -36.68 |
| Consensus MFE | -17.76 |
| Energy contribution | -17.97 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.529280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18803415 96 - 27905053 ------AAUCACCAGAUUUCCCGACAGCGACCACGAUGGUCGGGCAGAUAGCCGGGCUGGCAGGGGAAGUCGAUCAUGGCCACCAUCAUGGUCUUCUCGGUG ------...((((.((((((((..((((((((.....)))).(((.....)))..))))....))))))))((((((((......)))))))).....)))) ( -42.80, z-score = -3.12, R) >droSim1.chr3R 18616689 98 - 27517382 ----CGAAUCACCAGAUUUCCCGACAACGACCACGAUGGUCGGGCAGAUAGCCCGGCUGGCAGGGGAAGUCGAUCAUGGCCACCAUCAUGGGCUUCUCGGUG ----.....((((.((((((((.....((....))..((((((((.....)))))))).....))))))))((....((((.........))))..)))))) ( -37.20, z-score = -1.38, R) >droSec1.super_0 19167546 98 - 21120651 ----CGGAUCACCAGAUUUCCCGACAACGACCACGAUGGUCGGGCAGAUAGCCCGGCUGGCAGGGGAAGUCGAUCAUGGCCACCAUCAUGGUCUUCUCGGUG ----.....((((.((((((((.....((....))..((((((((.....)))))))).....))))))))((((((((......)))))))).....)))) ( -41.00, z-score = -2.27, R) >droYak2.chr3R 19663077 96 - 28832112 ------AAUCACCAGAUUUCCCGACAGCGACCACGAUGGUCGGGCAGAUAGCCCGGCUGGCAGGGAAAGUCGAUCAUGGCCACCAUCAUGGGCUUCUCGGUG ------...(((((((((((((..((((((((.....))))((((.....)))).))))...))))))(((.((.((((...)))).)).))).))).)))) ( -42.80, z-score = -3.29, R) >droEre2.scaffold_4820 1203635 96 + 10470090 ------AAUCACCAGAUUUCCCGACAGCGACCGCGAUGGUCGGGCAGAUAGCCCGGCUGGCAGGGGAAGUCGAUCAUGGCCACCAUCAUGGGCUUCUCGGUG ------...((((......(((..((((((((.....))))((((.....)))).))))...)))((((((.((.((((...)))).)).))))))..)))) ( -42.80, z-score = -2.53, R) >droAna3.scaffold_13340 11179480 102 + 23697760 UUUGCCGUUUACCAGAUUUCCCGGCACCGACCACGGUGGUCGGGCAGGUAGCCCGGCUGGCAGGGGAAGUCUAUCAUGGCCACCUCCAUGAUUUUCUCGGUA ..(((((......(((((((((.((((((....))))((((((((.....)))))))).))..)))))))))(((((((......))))))).....))))) ( -47.30, z-score = -3.37, R) >droWil1.scaffold_181089 11140380 94 + 12369635 --------CUACCAGAUUUCACGGCAACGGCCACGACUGUCGGGAAGAUAACCAGGCUGGCAAGGGAAAUCAAUCAUGGCUACCUGCAUAAUAUUUUUCUCA --------......(((.((..(((....)))..))..)))(((((((....((((.((((....((......))...))))))))........))))))). ( -21.10, z-score = -0.27, R) >droVir3.scaffold_12822 3991432 100 + 4096053 --AUCAGAUCACCAUAUCUCACGGCAGCGUCCACGGCUGUCAGGCAGGUAGCCUGGCUGACAGGGAAAAUCAAUCAUAGCAACCUCCAUAAUUUUUUCGGUU --........(((.........((((((.......)))))).((.((((..((((.....))))((.......))......))))))...........))). ( -23.70, z-score = -0.51, R) >droMoj3.scaffold_6540 1343659 100 + 34148556 --UGGGGUUUACCAUAUCUCGCGGCACCGUCCGCGACUGUCGGGCAGGUAGCCCGGCUGGCAGGGGAAGUCGAUCAUGGCCACCUCCAUGAUCUUCUCGGUG --.(((((.......)))))....((((((((.(..(((((((((.....))))))).))..).)))((..((((((((......))))))))..))))))) ( -40.90, z-score = -0.85, R) >droGri2.scaffold_15074 3608096 97 - 7742996 -----GAAUCACCAUAUCUCACGGCAGCGUCCACGACUGUCGGGCAAAUAGCCGGGCUGACAGGGAAAAUCAAUCAUGGCCACCUCCAUAAUUUUCUCGGUU -----.....(((....(((.((((((((....)).))))))(((.....))))))......((((((((.....((((......)))).))))))))))). ( -27.20, z-score = -1.37, R) >consensus ______AAUCACCAGAUUUCCCGACAGCGACCACGAUGGUCGGGCAGAUAGCCCGGCUGGCAGGGGAAGUCGAUCAUGGCCACCAUCAUGAUCUUCUCGGUG .........((((.........................(((((((.....)))))))......((((((......((((......))))...)))))))))) (-17.76 = -17.97 + 0.21)
| Location | 18,803,419 – 18,803,515 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 79.19 |
| Shannon entropy | 0.41606 |
| G+C content | 0.56439 |
| Mean single sequence MFE | -34.72 |
| Consensus MFE | -19.33 |
| Energy contribution | -18.44 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.571392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
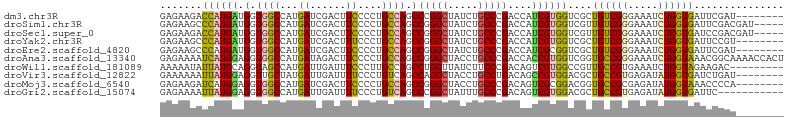
>dm3.chr3R 18803419 96 + 27905053 GAGAAGACCAUGAUGGUGGCCAUGAUCGACUUCCCCUGCCAGCCCGGCUAUCUGCCCGACCAUCGUGGUCGCUGUCGGGAAAUCUGGUGAUUCGAU-------- .....(((((((((((((((...((......))....))).....(((.....)))..))))))))))))...(((((...(((....))))))))-------- ( -32.90, z-score = -0.69, R) >droSim1.chr3R 18616693 99 + 27517382 GAGAAGCCCAUGAUGGUGGCCAUGAUCGACUUCCCCUGCCAGCCGGGCUAUCUGCCCGACCAUCGUGGUCGUUGUCGGGAAAUCUGGUGAUUCGACGAU----- .....(.(((((((((((((...((......))....)))...(((((.....))))))))))))))).)((((((((...(((....)))))))))))----- ( -38.90, z-score = -1.70, R) >droSec1.super_0 19167550 99 + 21120651 GAGAAGACCAUGAUGGUGGCCAUGAUCGACUUCCCCUGCCAGCCGGGCUAUCUGCCCGACCAUCGUGGUCGUUGUCGGGAAAUCUGGUGAUCCGACGAU----- .....(((((((((((((((...((......))....)))...(((((.....)))))))))))))))))((((((((...(((....)))))))))))----- ( -46.20, z-score = -3.81, R) >droYak2.chr3R 19663081 96 + 28832112 GAGAAGCCCAUGAUGGUGGCCAUGAUCGACUUUCCCUGCCAGCCGGGCUAUCUGCCCGACCAUCGUGGUCGCUGUCGGGAAAUCUGGUGAUUCCGU-------- ((..((((((((((((((((...((.......))...)))...(((((.....)))))))))))))))..))).)).((((.((....))))))..-------- ( -38.40, z-score = -1.86, R) >droEre2.scaffold_4820 1203639 96 - 10470090 GAGAAGCCCAUGAUGGUGGCCAUGAUCGACUUCCCCUGCCAGCCGGGCUAUCUGCCCGACCAUCGCGGUCGCUGUCGGGAAAUCUGGUGAUUCGAU-------- (((..(((((((........))))...((.(((((....((((.((((.....))))((((.....))))))))..))))).)).)))..)))...-------- ( -35.00, z-score = -0.58, R) >droAna3.scaffold_13340 11179484 104 - 23697760 GAGAAAAUCAUGGAGGUGGCCAUGAUAGACUUCCCCUGCCAGCCGGGCUACCUGCCCGACCACCGUGGUCGGUGCCGGGAAAUCUGGUAAACGGCAAAACCACU ..............(((.(((....((((.(((((..((..(((((((.....))))((((.....))))))))).))))).))))......)))...)))... ( -39.10, z-score = -1.52, R) >droWil1.scaffold_181089 11140384 95 - 12369635 AAAAAUAUUAUGCAGGUAGCCAUGAUUGAUUUCCCUUGCCAGCCUGGUUAUCUUCCCGACAGUCGUGGCCGUUGCCGUGAAAUCUGGUAGAAGAC--------- ..............((((((((((((((.........(((.....)))...........))))))))))...))))......(((......))).--------- ( -21.65, z-score = -0.18, R) >droVir3.scaffold_12822 3991436 96 - 4096053 GAAAAAAUUAUGGAGGUUGCUAUGAUUGAUUUUCCCUGUCAGCCAGGCUACCUGCCUGACAGCCGUGGACGCUGCCGUGAGAUAUGGUGAUCUGAU-------- .............((((..(((((........((((((((((.((((...)))).)))))))....)))(((....)))...)))))..))))...-------- ( -28.70, z-score = -1.80, R) >droMoj3.scaffold_6540 1343663 96 - 34148556 GAGAAGAUCAUGGAGGUGGCCAUGAUCGACUUCCCCUGCCAGCCGGGCUACCUGCCCGACAGUCGCGGACGGUGCCGCGAGAUAUGGUAAACCCCA-------- (((..((((((((......))))))))..)))....((((((.(((((.....))))).)..((((((......))))))....))))).......-------- ( -39.50, z-score = -2.34, R) >droGri2.scaffold_15074 3608100 93 + 7742996 GAGAAAAUUAUGGAGGUGGCCAUGAUUGAUUUUCCCUGUCAGCCCGGCUAUUUGCCCGACAGUCGUGGACGCUGCCGUGAGAUAUGGUGAUUC----------- .......((((((.((((.(((((((((.........(....)..(((.....)))...))))))))).)))).)))))).............----------- ( -26.80, z-score = -0.69, R) >consensus GAGAAGACCAUGAAGGUGGCCAUGAUCGACUUCCCCUGCCAGCCGGGCUAUCUGCCCGACCAUCGUGGUCGCUGCCGGGAAAUCUGGUGAUUCGAU________ .......((((((.(.((((...((......))....)))).)(((((.....)))))....))))))....((((((.....))))))............... (-19.33 = -18.44 + -0.89)
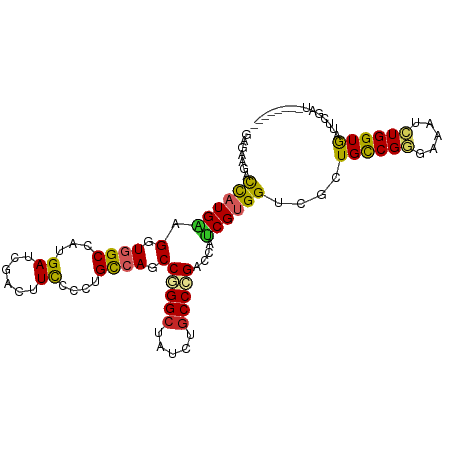
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:36:54 2011