
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,777,591 – 18,777,727 |
| Length | 136 |
| Max. P | 0.657987 |

| Location | 18,777,591 – 18,777,688 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 67.66 |
| Shannon entropy | 0.62299 |
| G+C content | 0.39788 |
| Mean single sequence MFE | -24.47 |
| Consensus MFE | -8.84 |
| Energy contribution | -9.47 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.657987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
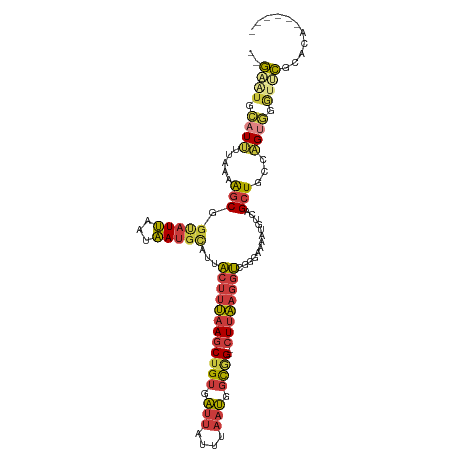
>dm3.chr3R 18777591 97 + 27905053 ---UCAACUGAGCACACUCAUUACAGACGCGAAUGCAUUUUAAAAGCGGUAUUAAUAAUGCAUUAC------UUUAAGCUGUGAUUAU--UUAAUGGCGGCUUAAGGU------- ---.....((((....))))...........((((((((((((........)))).))))))))((------((((((((((.(((..--..))).))))))))))))------- ( -25.90, z-score = -2.85, R) >droWil1.scaffold_181089 11114037 115 - 12369635 UUAACUUGGCAGCAGAUCCUUUGCCAGUUGGCACACACCCGAAAAUUUUUAUGAAUGGCAGCUUAAAAUAUUUGCAUUCUGCAACUGGGGCCAAUAACCGGUAGAACUUUUACAU .....(((((..(((.....((((((.((((.......)))).............))))))..........((((.....)))))))..)))))......(((((...))))).. ( -22.24, z-score = 0.91, R) >dp4.chr2 24685652 79 - 30794189 ----------------CCCACU--GAGCACACUCUCAUUACGCGUGCGGCAUCCAUAAUGCAACGCG----CUUUAAGCUGUGAUUAU--UUAAUGGCCGGCU------------ ----------------.....(--(((......))))....(((((..((((.....))))..))))----)....((((((.(((..--..))).).)))))------------ ( -19.80, z-score = -0.22, R) >droAna3.scaffold_13340 11153875 97 - 23697760 ---GCAAGUGGAGCCACUCAUUAUGGGCGCGUAGGCAUUUUAAAAGCCUUAUUAAUAAUGCAUUAC------UUUAAGCCGUGAUUAU--UUAAUGGCAGCUUAAGGU------- ---(((((((....)))).((((.((((...(((.....)))...))))...))))..)))...((------(((((((.((.(((..--..))).)).)))))))))------- ( -25.10, z-score = -0.70, R) >droEre2.scaffold_4820 1178722 97 - 10470090 ---UCAACUGAGCACACUCAUUACAGACGCGAGUGCAUUUCAAAAGCGGCAUUAAUAAUGCAUUAC------UUUAAGCUGUGAUUAU--UUAAUGGCGGCUUAAGGU------- ---.....((((....)))).......(((...((.....))...)))((((.....))))...((------((((((((((.(((..--..))).))))))))))))------- ( -25.90, z-score = -2.20, R) >droYak2.chr3R 19635493 97 + 28832112 ---UCAACUGAGCACACUCAUUACAGACGCGAAUGCAUUUCGAGAGCGGCAUUAAUAAUGCAUUAC------UUUAAGCUGUGAUUAU--UUAAUGGCGGCUUAAGGU------- ---.....((((....))))...........(((((((((((....))).......))))))))((------((((((((((.(((..--..))).))))))))))))------- ( -25.01, z-score = -1.83, R) >droSec1.super_0 19141919 97 + 21120651 ---UCAACUGAGCACACUCAUUACAGACGCGAAUGCAUUUUAAAAGCGGUAUUAAUAAUGCAUUAC------UUUAAGCUGUGAUUAU--UUAAUGGCGGCUUAAGGU------- ---.....((((....))))...........((((((((((((........)))).))))))))((------((((((((((.(((..--..))).))))))))))))------- ( -25.90, z-score = -2.85, R) >droSim1.chr3R 18588872 97 + 27517382 ---UCAACUGAGCACACUCAUUACAGACGCGAAUGCAUUUUAAAAGCGGUAUUAAUAAUGCAUUAC------UUUAAGCUGUGAUUAU--UUAAUGGCGGCUUAAGGU------- ---.....((((....))))...........((((((((((((........)))).))))))))((------((((((((((.(((..--..))).))))))))))))------- ( -25.90, z-score = -2.85, R) >consensus ___UCAACUGAGCACACUCAUUACAGACGCGAAUGCAUUUUAAAAGCGGUAUUAAUAAUGCAUUAC______UUUAAGCUGUGAUUAU__UUAAUGGCGGCUUAAGGU_______ ..................................(((((.................)))))...........(((((((.((.((((....)))).)).)))))))......... ( -8.84 = -9.47 + 0.63)



| Location | 18,777,618 – 18,777,727 |
|---|---|
| Length | 109 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 74.61 |
| Shannon entropy | 0.49127 |
| G+C content | 0.39699 |
| Mean single sequence MFE | -29.34 |
| Consensus MFE | -12.12 |
| Energy contribution | -12.69 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.546021 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 18777618 109 + 27905053 --GAAUGCAUUUUAAAAGCGGUAUUAAUAAUGCAUUACUUUAAGCUGUGAUUAUUUAAUGGCGG--CUUAAGGUCGGGAAAAUGUCAGCUGCCAGUGUGCUCGCACACACACA --.((((((((((((........)))).))))))))((((((((((((.(((....))).))))--)))))))).((.(..........).)).(((((....)))))..... ( -31.30, z-score = -1.73, R) >droWil1.scaffold_181089 11114077 107 - 12369635 GAAAAUUUUUAUGAAUGGCAGCUUAAAAUAUUUGCAUUCUGCAACUGGGGCCAAUAACCGGUAGAACUUUUACAUGGGUAAAAG-CAGAUAGAUGCAAAUAUUUAAUU----- .........................(((((((((((((((((.(((((.........))))).....((((((....)))))))-))))...))))))))))))....----- ( -27.30, z-score = -2.86, R) >droAna3.scaffold_13340 11153902 103 - 23697760 --GUAGGCAUUUUAAAAGCCUUAUUAAUAAUGCAUUACUUUAAGCCGUGAUUAUUUAAUGGCAG--CUUAAGGUCAGAAAAAUGUCAGCUGCCAGCCAGCGCGCAUA------ --((.((((((((....((.(((....))).))...(((((((((.((.(((....))).)).)--))))))))....)))))))).((((.....)))))).....------ ( -26.20, z-score = -0.89, R) >droEre2.scaffold_4820 1178749 103 - 10470090 --GAGUGCAUUUCAAAAGCGGCAUUAAUAAUGCAUUACUUUAAGCUGUGAUUAUUUAAUGGCGG--CUUAAGGUCGGAAAAAUGUCAGCUGCCAGUGGGUGCGCACA------ --..(((((((.((...(((((..............((((((((((((.(((....))).))))--))))))))..((......)).)))))...)))))))))...------ ( -34.60, z-score = -2.70, R) >droYak2.chr3R 19635520 103 + 28832112 --GAAUGCAUUUCGAGAGCGGCAUUAAUAAUGCAUUACUUUAAGCUGUGAUUAUUUAAUGGCGG--CUUAAGGUCGGGAAAAUGUCAGCUGCCAGUGGGUUCGCACA------ --((((.((((..(..(((((((((.....((....((((((((((((.(((....))).))))--))))))))))....)))))).))).).)))).)))).....------ ( -30.30, z-score = -1.48, R) >droSec1.super_0 19141946 103 + 21120651 --GAAUGCAUUUUAAAAGCGGUAUUAAUAAUGCAUUACUUUAAGCUGUGAUUAUUUAAUGGCGG--CUUAAGGUCGGGAAAAUUUCAGCUGCCAGUGGGUUCGCACA------ --((((.((((.....(((.((((.....))))...((((((((((((.(((....))).))))--)))))))).............)))...)))).)))).....------ ( -28.30, z-score = -1.88, R) >droSim1.chr3R 18588899 108 + 27517382 --GAAUGCAUUUUAAAAGCGGUAUUAAUAAUGCAUUACUUUAAGCUGUGAUUAUUUAAUGGCGG--CUUAAGGUCGGGAAAAUGUCAGCUGCC-GUGGGUUCGCACACACACA --.((((((((((((........)))).))))))))((((((((((((.(((....))).))))--)))))))).(.(((.(((.(....).)-))...))).)......... ( -27.40, z-score = -0.90, R) >consensus __GAAUGCAUUUUAAAAGCGGUAUUAAUAAUGCAUUACUUUAAGCUGUGAUUAUUUAAUGGCGG__CUUAAGGUCGGGAAAAUGUCAGCUGCCAGUGGGUUCGCACA______ ..((((.((((.....(((.(((((...)))))...((((((((((((.(((....))).))))..)))))))).............)))...)))).))))........... (-12.12 = -12.69 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:36:49 2011