
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,364,601 – 8,364,697 |
| Length | 96 |
| Max. P | 0.927598 |

| Location | 8,364,601 – 8,364,697 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 81.08 |
| Shannon entropy | 0.37081 |
| G+C content | 0.42477 |
| Mean single sequence MFE | -24.02 |
| Consensus MFE | -15.90 |
| Energy contribution | -16.68 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.641864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
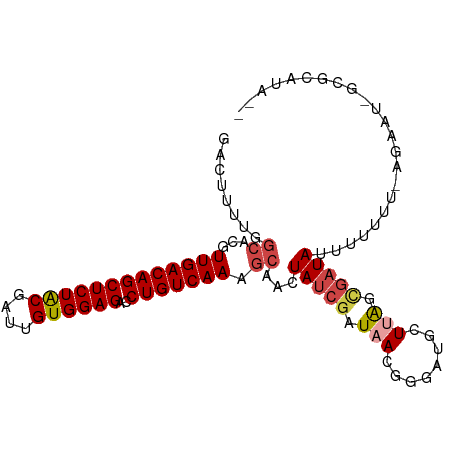
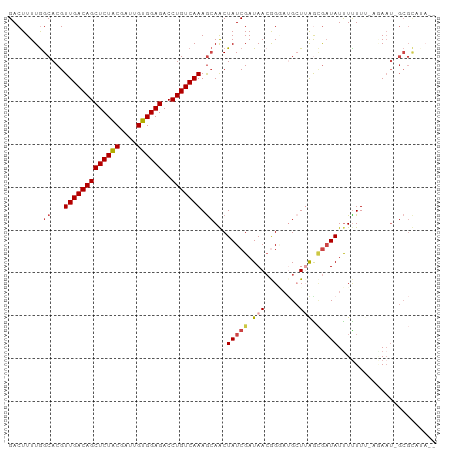
>dm3.chr2L 8364601 96 + 23011544 GACUUUUGGCACGUUGACAGCUCUACGAU-GUGGAGACCUGUCAAAGCAGCUAUCGAUAACGGGAUGCUUAGCGAUAUUUUUU--UUAAU-GCGCUUACA ........((...(((((((((((((...-))))))..))))))).))(((..(((....)))...))).((((.((((....--..)))-))))).... ( -24.30, z-score = -1.17, R) >droAna3.scaffold_12916 5449642 86 + 16180835 GACUUUUAGCACGUUGACAGCUCUGCUAUUGUGGAGACCUGUCAAAA-AACUAUCGAUAACAG--CGAUUGGUAUUAUCUUUUCAAGAA----------- ......(((....((((((((((..(....)..)))..)))))))..-..)))..(((((..(--(.....)).)))))..........----------- ( -18.20, z-score = -0.92, R) >droYak2.chr2L 11002643 98 + 22324452 GACUUUUGGCACGUUGACAGCUCUACGAUUGUGGAGACCUGUCAAAGCAACUAUCGAUAAGGGGAGGGUAAGCGAUAUUUUUUUGUAAAUCGCGUACA-- ..(((((.((...(((((((((((((....))))))..))))))).))..((........)))))))(((.(((((............))))).))).-- ( -25.40, z-score = -1.11, R) >droEre2.scaffold_4929 17273924 95 + 26641161 GACUUUUGGCACGUUGACAGCUCUACGAUUGUGGAGACCUGUCAAAGCAACUAUCGAUAACAGAAUGCUAAGCGAUAUUUCUUUUCUAAU-GCGUG---- .....((((((..(((((((((((((....))))))..)))))))........((.......)).))))))(((.((((........)))-)))).---- ( -22.10, z-score = -0.95, R) >droSec1.super_3 3860000 99 + 7220098 GACUUUUGGCACGUUGACAGCUCUACGAUUGUGGAGACCUGUCAAAGCAGCUAUCGAUAACGGCAUGCUUAAAGAUAUUUUUUUUGGAAU-GCGCAUAAC ......((((...(((((((((((((....))))))..)))))))....)))).........((((..(((((((.....))))))).))-))....... ( -25.70, z-score = -1.67, R) >droSim1.chr2L 8150158 97 + 22036055 GACUUUUGGCACGUUGACAGCUCUACGAUUGUGGAGACCUGUCAAAGCAGCUAUCGAUAACGGGAUGCUUAACGAUAUUUUUU--AGACU-GCGCUUACA .......(((...(((((((((((((....))))))..))))))).(((((((..((((.((..(....)..)).))))...)--)).))-))))).... ( -28.40, z-score = -2.95, R) >consensus GACUUUUGGCACGUUGACAGCUCUACGAUUGUGGAGACCUGUCAAAGCAACUAUCGAUAACGGGAUGCUUAGCGAUAUUUUUUU_AGAAU_GCGCAUA__ ........((...(((((((((((((....))))))..))))))).))...(((((.(((........))).)))))....................... (-15.90 = -16.68 + 0.78)

| Location | 8,364,601 – 8,364,697 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 81.08 |
| Shannon entropy | 0.37081 |
| G+C content | 0.42477 |
| Mean single sequence MFE | -22.50 |
| Consensus MFE | -14.41 |
| Energy contribution | -15.05 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.927598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
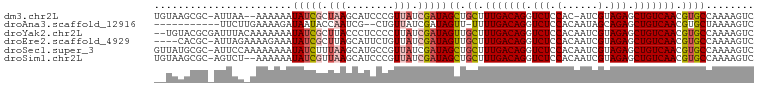
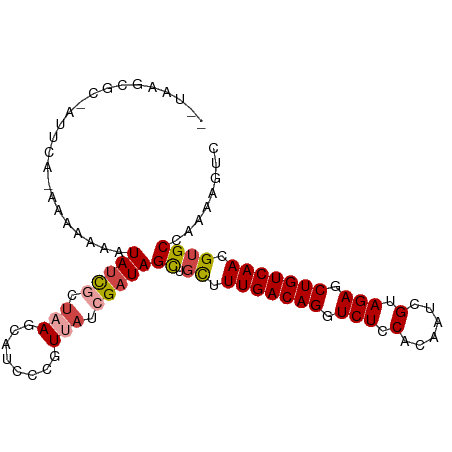
>dm3.chr2L 8364601 96 - 23011544 UGUAAGCGC-AUUAA--AAAAAAUAUCGCUAAGCAUCCCGUUAUCGAUAGCUGCUUUGACAGGUCUCCAC-AUCGUAGAGCUGUCAACGUGCCAAAAGUC .....((((-.....--..........((..((((((........))).))))).(((((((.(((.(..-...).))).))))))).))))........ ( -22.20, z-score = -1.75, R) >droAna3.scaffold_12916 5449642 86 - 16180835 -----------UUCUUGAAAAGAUAAUACCAAUCG--CUGUUAUCGAUAGUU-UUUUGACAGGUCUCCACAAUAGCAGAGCUGUCAACGUGCUAAAAGUC -----------..........(((((((.(....)--.)))))))..((((.-..(((((((.(((.(......).))).)))))))...))))...... ( -20.90, z-score = -2.23, R) >droYak2.chr2L 11002643 98 - 22324452 --UGUACGCGAUUUACAAAAAAAUAUCGCUUACCCUCCCCUUAUCGAUAGUUGCUUUGACAGGUCUCCACAAUCGUAGAGCUGUCAACGUGCCAAAAGUC --.(((.(((((............))))).)))................(..((.(((((((.(((.(......).))).))))))).))..)....... ( -21.80, z-score = -2.35, R) >droEre2.scaffold_4929 17273924 95 - 26641161 ----CACGC-AUUAGAAAAGAAAUAUCGCUUAGCAUUCUGUUAUCGAUAGUUGCUUUGACAGGUCUCCACAAUCGUAGAGCUGUCAACGUGCCAAAAGUC ----((((.-......((((...(((((..((((.....)))).)))))....))))(((((.(((.(......).))).)))))..))))......... ( -23.30, z-score = -1.75, R) >droSec1.super_3 3860000 99 - 7220098 GUUAUGCGC-AUUCCAAAAAAAAUAUCUUUAAGCAUGCCGUUAUCGAUAGCUGCUUUGACAGGUCUCCACAAUCGUAGAGCUGUCAACGUGCCAAAAGUC .....((((-....................(((((.((..((...))..)))))))((((((.(((.(......).))).))))))..))))........ ( -23.00, z-score = -1.89, R) >droSim1.chr2L 8150158 97 - 22036055 UGUAAGCGC-AGUCU--AAAAAAUAUCGUUAAGCAUCCCGUUAUCGAUAGCUGCUUUGACAGGUCUCCACAAUCGUAGAGCUGUCAACGUGCCAAAAGUC .....((((-(((..--......(((((...(((.....)))..)))))))))).(((((((.(((.(......).))).)))))))...))........ ( -23.80, z-score = -2.38, R) >consensus __UAAGCGC_AUUCA_AAAAAAAUAUCGCUAAGCAUCCCGUUAUCGAUAGCUGCUUUGACAGGUCUCCACAAUCGUAGAGCUGUCAACGUGCCAAAAGUC .......................(((((.(((........))).)))))((.((.(((((((.(((.(......).))).))))))).))))........ (-14.41 = -15.05 + 0.64)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:25:48 2011