| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,771,833 – 18,771,885 |
| Length | 52 |
| Max. P | 0.602894 |

| Location | 18,771,833 – 18,771,885 |
|---|---|
| Length | 52 |
| Sequences | 7 |
| Columns | 63 |
| Reading direction | forward |
| Mean pairwise identity | 63.11 |
| Shannon entropy | 0.68051 |
| G+C content | 0.48188 |
| Mean single sequence MFE | -12.49 |
| Consensus MFE | -5.96 |
| Energy contribution | -6.61 |
| Covariance contribution | 0.66 |
| Combinations/Pair | 1.55 |
| Mean z-score | -0.63 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.602894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
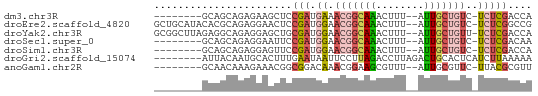
>dm3.chr3R 18771833 52 + 27905053 UGGUCGAGA-GACAGCAAU--AAAGUUUGCCGUUUCAUCGGAGCUUCUCUGCUGC-------- ...((((((-(((.((((.--.....)))).))))).))))(((......)))..-------- ( -16.00, z-score = -1.37, R) >droEre2.scaffold_4820 1173154 60 - 10470090 CGGCCGAGA-GACAGCAAU--AAAGUUUGCCGUUCCAUCGGAGUUCCUCUGCGUGUAUGCAGC ...(((((.-.((.((((.--.....)))).))..).)))).......(((((....))))). ( -16.80, z-score = -0.97, R) >droYak2.chr3R 19629432 60 + 28832112 UGGUCGAGA-AACAGCAAU--AAAGUUUGCCGUUCCAUCGCAGCUCCUCUGCCUCUAAGCCGC .(((..(((-(((.((((.--.....)))).))).....((((.....)))).)))..))).. ( -14.80, z-score = -1.29, R) >droSec1.super_0 19136155 52 + 21120651 UUGUCGAGA-GACAGCAAU--AAAGUUUGCCGUUCCAUCGGAAUUCCUCUGCUGC-------- .((((....-))))((((.--.....)))).....((.((((.....)))).)).-------- ( -11.40, z-score = -0.70, R) >droSim1.chr3R 18583026 52 + 27517382 UGGUCGAGA-GACAGCAAU--AAAGUUUGCCGUUCCAUCGGAACUCCUCUGCUGC-------- .(((.(((.-((..((((.--.....)))).(((((...)))))))))).)))..-------- ( -13.50, z-score = -0.90, R) >droGri2.scaffold_15074 3578494 55 + 7742996 UUUUUAAGAUGAGUGCAGUCUAAGGUCUAAGGAAUUAUUCAAAGUGCAUUGUAAU-------- ...........((((((.(......((....)).........).)))))).....-------- ( -4.46, z-score = 1.20, R) >anoGam1.chr2R 43084342 52 - 62725911 AACGCGUAA-GAACGCAAU--AAACGCUUCCGUUUGUCCGCCGUUUCUUUGUUGC-------- ...(((.((-((((((.((--(((((....)))))))..).)).)))))...)))-------- ( -10.50, z-score = -0.39, R) >consensus UGGUCGAGA_GACAGCAAU__AAAGUUUGCCGUUCCAUCGGAGCUCCUCUGCUGC________ ...((((...(((.((((........)))).)))...))))...................... ( -5.96 = -6.61 + 0.66)



| Location | 18,771,833 – 18,771,885 |
|---|---|
| Length | 52 |
| Sequences | 7 |
| Columns | 63 |
| Reading direction | reverse |
| Mean pairwise identity | 63.11 |
| Shannon entropy | 0.68051 |
| G+C content | 0.48188 |
| Mean single sequence MFE | -14.42 |
| Consensus MFE | -4.67 |
| Energy contribution | -5.29 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.523209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
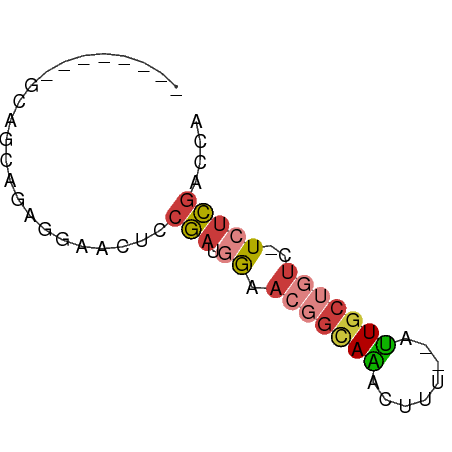
>dm3.chr3R 18771833 52 - 27905053 --------GCAGCAGAGAAGCUCCGAUGAAACGGCAAACUUU--AUUGCUGUC-UCUCGACCA --------..(((......))).(((.((.(((((((.....--.))))))).-))))).... ( -13.70, z-score = -1.29, R) >droEre2.scaffold_4820 1173154 60 + 10470090 GCUGCAUACACGCAGAGGAACUCCGAUGGAACGGCAAACUUU--AUUGCUGUC-UCUCGGCCG .((((......)))).((....((((.(..(((((((.....--.))))))).-.))))))). ( -21.10, z-score = -3.15, R) >droYak2.chr3R 19629432 60 - 28832112 GCGGCUUAGAGGCAGAGGAGCUGCGAUGGAACGGCAAACUUU--AUUGCUGUU-UCUCGACCA (((((((..........)))))))((.((((((((((.....--.))))))))-))))..... ( -22.90, z-score = -2.62, R) >droSec1.super_0 19136155 52 - 21120651 --------GCAGCAGAGGAAUUCCGAUGGAACGGCAAACUUU--AUUGCUGUC-UCUCGACAA --------...............(((.(..(((((((.....--.))))))).-.)))).... ( -12.60, z-score = -0.96, R) >droSim1.chr3R 18583026 52 - 27517382 --------GCAGCAGAGGAGUUCCGAUGGAACGGCAAACUUU--AUUGCUGUC-UCUCGACCA --------(((((((..((((((((......)))..))))).--.))))))).-......... ( -16.00, z-score = -1.52, R) >droGri2.scaffold_15074 3578494 55 - 7742996 --------AUUACAAUGCACUUUGAAUAAUUCCUUAGACCUUAGACUGCACUCAUCUUAAAAA --------.......((((.(((((...............))))).))))............. ( -3.96, z-score = -0.69, R) >anoGam1.chr2R 43084342 52 + 62725911 --------GCAACAAAGAAACGGCGGACAAACGGAAGCGUUU--AUUGCGUUC-UUACGCGUU --------((....(((((...((((..(((((....)))))--.)))).)))-))..))... ( -10.70, z-score = 0.14, R) >consensus ________GCAGCAGAGGAACUCCGAUGGAACGGCAAACUUU__AUUGCUGUC_UCUCGACCA .......................(((....(((((((........)))))))....))).... ( -4.67 = -5.29 + 0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:36:46 2011