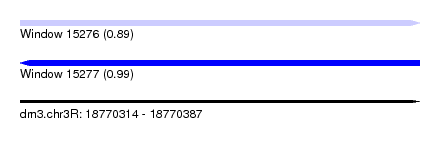
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,770,314 – 18,770,387 |
| Length | 73 |
| Max. P | 0.990690 |

| Location | 18,770,314 – 18,770,387 |
|---|---|
| Length | 73 |
| Sequences | 6 |
| Columns | 80 |
| Reading direction | forward |
| Mean pairwise identity | 73.87 |
| Shannon entropy | 0.49582 |
| G+C content | 0.52060 |
| Mean single sequence MFE | -17.02 |
| Consensus MFE | -8.73 |
| Energy contribution | -8.70 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.894345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
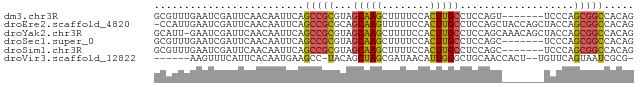
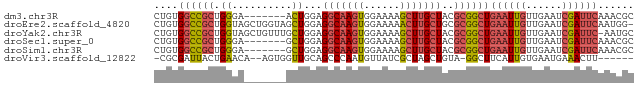
>dm3.chr3R 18770314 73 + 27905053 GCGUUUGAAUCGAUUCAACAAUUCAGCCGCGUAGCAAGCUUUUCCACUUGCCUCCAGU-------UCCCAGCGGCCACAG .....(((((.(......).)))))((((((..(((((........)))))..)..(.-------...).)))))..... ( -15.00, z-score = -1.17, R) >droEre2.scaffold_4820 1171797 79 - 10470090 -CCAUUGAAUCGAUUCAACAAUUCAGCCGCGCAGCAAGUUUUUCCACUUGCCUCCAGCUACCAGCUACCAGCGGCCACAG -....(((((.(......).)))))(((((...((((((......))))))....(((.....)))....)))))..... ( -20.90, z-score = -3.89, R) >droYak2.chr3R 19628271 79 + 28832112 GCAUU-GAAUCGAUUCAACAAUUCAGCCGCGUAGCAAGCUUUUCCACUUGCCUCCAGCAAACAGCUACCAGCGGCCACAG ....(-((((.(......).)))))((((((..(((((........)))))..).(((.....)))....)))))..... ( -19.60, z-score = -2.60, R) >droSec1.super_0 19134670 73 + 21120651 GCGUUUGAAUCGAUUCAACAAUUCAGCCGCGUAGCAAGCUUUUCCACUUGCCUCCAGC-------UCCCAGCGGCCACAG (((.((((((.(......).)))))).)))((.(((((........)))))..((.((-------.....))))..)).. ( -15.10, z-score = -0.79, R) >droSim1.chr3R 18581524 73 + 27517382 GCGUUUGAAUCGAUUCAACAAUUCAGCCGCGUAGCAAGCUUUUCCACUUGCCUCCAGC-------UCCCAGCGGCCACAG (((.((((((.(......).)))))).)))((.(((((........)))))..((.((-------.....))))..)).. ( -15.10, z-score = -0.79, R) >droVir3.scaffold_12822 3968317 70 - 4096053 ------AAGUUUCAUUCACAAUGAAGCC-UACAGCUAGCGAUAACAUUGGGCUGCAACCACU--UGUUCAGUAAUCGCG- ------..(((((((.....))))))).-........(((((...(((((((..........--.))))))).))))).- ( -16.40, z-score = -1.50, R) >consensus GCGUUUGAAUCGAUUCAACAAUUCAGCCGCGUAGCAAGCUUUUCCACUUGCCUCCAGC_______UCCCAGCGGCCACAG .........................(((((...(((((........)))))...................)))))..... ( -8.73 = -8.70 + -0.03)
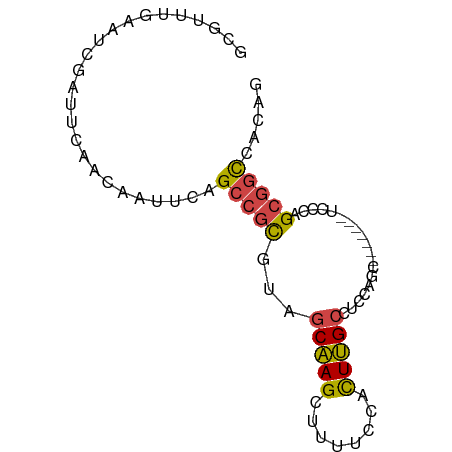
| Location | 18,770,314 – 18,770,387 |
|---|---|
| Length | 73 |
| Sequences | 6 |
| Columns | 80 |
| Reading direction | reverse |
| Mean pairwise identity | 73.87 |
| Shannon entropy | 0.49582 |
| G+C content | 0.52060 |
| Mean single sequence MFE | -25.48 |
| Consensus MFE | -15.71 |
| Energy contribution | -16.55 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.990690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
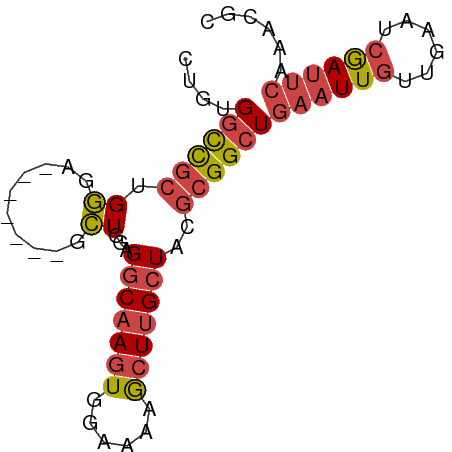
>dm3.chr3R 18770314 73 - 27905053 CUGUGGCCGCUGGGA-------ACUGGAGGCAAGUGGAAAAGCUUGCUACGCGGCUGAAUUGUUGAAUCGAUUCAAACGC ..((((((((.(...-------.)..(.(((((((......))))))).))))))(((((((......)))))))..))) ( -26.00, z-score = -2.74, R) >droEre2.scaffold_4820 1171797 79 + 10470090 CUGUGGCCGCUGGUAGCUGGUAGCUGGAGGCAAGUGGAAAAACUUGCUGCGCGGCUGAAUUGUUGAAUCGAUUCAAUGG- ....((((((...((((.....))))(.(((((((......))))))).)))))))((((((......)))))).....- ( -29.30, z-score = -2.79, R) >droYak2.chr3R 19628271 79 - 28832112 CUGUGGCCGCUGGUAGCUGUUUGCUGGAGGCAAGUGGAAAAGCUUGCUACGCGGCUGAAUUGUUGAAUCGAUUC-AAUGC ....((((((...((((.....))))(.(((((((......))))))).)))))))((((((......))))))-..... ( -29.30, z-score = -2.60, R) >droSec1.super_0 19134670 73 - 21120651 CUGUGGCCGCUGGGA-------GCUGGAGGCAAGUGGAAAAGCUUGCUACGCGGCUGAAUUGUUGAAUCGAUUCAAACGC ..((((((((.(...-------.)..(.(((((((......))))))).))))))(((((((......)))))))..))) ( -25.00, z-score = -1.74, R) >droSim1.chr3R 18581524 73 - 27517382 CUGUGGCCGCUGGGA-------GCUGGAGGCAAGUGGAAAAGCUUGCUACGCGGCUGAAUUGUUGAAUCGAUUCAAACGC ..((((((((.(...-------.)..(.(((((((......))))))).))))))(((((((......)))))))..))) ( -25.00, z-score = -1.74, R) >droVir3.scaffold_12822 3968317 70 + 4096053 -CGCGAUUACUGAACA--AGUGGUUGCAGCCCAAUGUUAUCGCUAGCUGUA-GGCUUCAUUGUGAAUGAAACUU------ -.(((((((((.....--))))))))).(((....((((....))))....-)))(((((.....)))))....------ ( -18.30, z-score = -1.13, R) >consensus CUGUGGCCGCUGGGA_______GCUGGAGGCAAGUGGAAAAGCUUGCUACGCGGCUGAAUUGUUGAAUCGAUUCAAACGC ....((((((.((..........))...(((((((......)))))))..))))))((((((......))))))...... (-15.71 = -16.55 + 0.84)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:36:44 2011