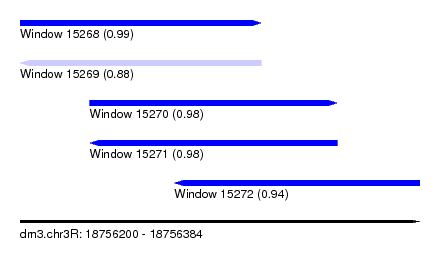
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,756,200 – 18,756,384 |
| Length | 184 |
| Max. P | 0.985638 |

| Location | 18,756,200 – 18,756,311 |
|---|---|
| Length | 111 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 71.26 |
| Shannon entropy | 0.54996 |
| G+C content | 0.50289 |
| Mean single sequence MFE | -37.23 |
| Consensus MFE | -20.27 |
| Energy contribution | -21.44 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.985638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
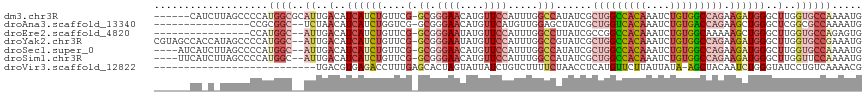
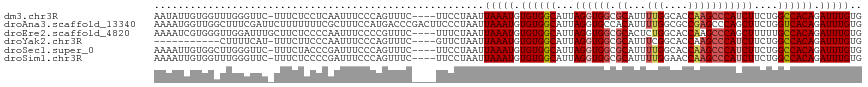
>dm3.chr3R 18756200 111 + 27905053 ------CAUCUUAGCCCCAUGGCGCAUUGACAUCAUCUGUUCG-GCGGGAACAUGUUCCAUUUGGCCAUAUCGCUGGCCACAAAUCUGUGGCCAGAAGAUGGGCUUGGUGCCAAAAUG ------((((..(((((.(((((((...((((.....))))..-)).((((....)))).....)))))(((.(((((((((....)))))))))..)))))))).))))........ ( -44.90, z-score = -2.84, R) >droAna3.scaffold_13340 11131689 99 - 23697760 ----------------CCGCGGC--UCUAACAUCAUCUGGUCG-GCGGGAACAUGUUCAUGUUGGAGCUAUCGCUGGUCACAAAUCUGUGACCAGAAGCUGGGCUCGGCGCCAAAAUG ----------------..(((((--((((((((...(((....-.)))(((....))))))))))))))....(((((((((....)))))))))..))..(((.....)))...... ( -37.90, z-score = -2.61, R) >droEre2.scaffold_4820 1157805 99 - 10470090 ----------------CCAUGGC--AUUGACAUCAUCUGUUCG-GCGGGAAUAUGUUCCAUUUGGCCUUAUCGCCGGCCACAAAUCUGUGGCAAAAAGCUGGGCUUGGUGCCAGAGUG ----------------...((((--((..(........(((((-((.((((....))))...((((......))))((((((....)))))).....))))))))..))))))..... ( -36.00, z-score = -1.52, R) >droYak2.chr3R 19613556 115 + 28832112 CGUAGCCACCAUAGCCCCAUGGC--AUUGACAUCAUCUGUUCG-GCGGGAAUAUGUUCCAUUUGGCCGUAUCGCUGGCCACAAAUCUGUGGCCAGAAGAUGGGCUUGGUGCCGAAAUG ....(.(((((.(((((.(((((--...(((((..((((....-.))))...))))).......)))))(((.(((((((((....)))))))))..))))))))))))).)...... ( -44.80, z-score = -2.04, R) >droSec1.super_0 19121012 111 + 21120651 ----AUCAUCUUAGCCCCAUGGC--AUUGACAUCAUCUGUUCG-GCGGGAACAUGUUCCAUUUGGCCAUAUCGCUGGCCACAAAUCUGUGGCCAGAAGAUGGGCUUGGUGCCAAAAUG ----...............((((--((..(..((((((....(-((.((((....)))).....)))......(((((((((....))))))))).))))))..)..))))))..... ( -43.80, z-score = -2.79, R) >droSim1.chr3R 18567263 111 + 27517382 ----UUCAUCUUAGCCCCAUGGC--AUUGACAUCAUCUGUUCG-GCGGGAACAUGUUCCAUUUGGCCAUAUCGCUGGCCACAAAUCUGUGGCCAGAAGAUGGGCUUGGUUCCAAAAUG ----.....((.(((((.(((((--...((((.....))))..-...((((....)))).....)))))(((.(((((((((....)))))))))..)))))))).)).......... ( -38.90, z-score = -1.91, R) >droVir3.scaffold_12822 3957468 90 - 4096053 ---------------------------UGACGUGAGACCUUUGAGCACUAGUAUUAUCUGUCUUUUCUAACCUCAUGUUCUUAUUAUA-AGCUACAAUCUGGGUAUCCUGUCAAAACG ---------------------------(((((.((.((((((((((..(((((..((.((.............)).))...)))))..-.))).)))...)))).)).)))))..... ( -14.32, z-score = -0.17, R) >consensus ______CA_C_UAGCCCCAUGGC__AUUGACAUCAUCUGUUCG_GCGGGAACAUGUUCCAUUUGGCCAUAUCGCUGGCCACAAAUCUGUGGCCAGAAGAUGGGCUUGGUGCCAAAAUG ..........................(((.(((((..(((((......)))))..........((((......(((((((((....)))))))))......))))))))).))).... (-20.27 = -21.44 + 1.17)


| Location | 18,756,200 – 18,756,311 |
|---|---|
| Length | 111 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 71.26 |
| Shannon entropy | 0.54996 |
| G+C content | 0.50289 |
| Mean single sequence MFE | -33.66 |
| Consensus MFE | -17.40 |
| Energy contribution | -18.43 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.876900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18756200 111 - 27905053 CAUUUUGGCACCAAGCCCAUCUUCUGGCCACAGAUUUGUGGCCAGCGAUAUGGCCAAAUGGAACAUGUUCCCGC-CGAACAGAUGAUGUCAAUGCGCCAUGGGGCUAAGAUG------ (((((((((.((..(((.(((..((((((((......)))))))).)))..)))...((((..(((((((....-.)))).(((...))).)))..)))))).)))))))))------ ( -41.60, z-score = -2.31, R) >droAna3.scaffold_13340 11131689 99 + 23697760 CAUUUUGGCGCCGAGCCCAGCUUCUGGUCACAGAUUUGUGACCAGCGAUAGCUCCAACAUGAACAUGUUCCCGC-CGACCAGAUGAUGUUAGA--GCCGCGG---------------- ......(((.....)))..((..((((((((......)))))))).....((((.(((((.(...((.((....-.)).))..).))))).))--)).))..---------------- ( -30.20, z-score = -1.70, R) >droEre2.scaffold_4820 1157805 99 + 10470090 CACUCUGGCACCAAGCCCAGCUUUUUGCCACAGAUUUGUGGCCGGCGAUAAGGCCAAAUGGAACAUAUUCCCGC-CGAACAGAUGAUGUCAAU--GCCAUGG---------------- ..((.(((((..((((...))))..))))).))..(..(((((((((..((..((....))......))..)))-)).(((.....)))....--))))..)---------------- ( -27.00, z-score = -0.24, R) >droYak2.chr3R 19613556 115 - 28832112 CAUUUCGGCACCAAGCCCAUCUUCUGGCCACAGAUUUGUGGCCAGCGAUACGGCCAAAUGGAACAUAUUCCCGC-CGAACAGAUGAUGUCAAU--GCCAUGGGGCUAUGGUGGCUACG ......(.(((((((((((((..((((((((......)))))))).))).((((.....((((....)))).))-))................--......))))).))))).).... ( -44.10, z-score = -2.55, R) >droSec1.super_0 19121012 111 - 21120651 CAUUUUGGCACCAAGCCCAUCUUCUGGCCACAGAUUUGUGGCCAGCGAUAUGGCCAAAUGGAACAUGUUCCCGC-CGAACAGAUGAUGUCAAU--GCCAUGGGGCUAAGAUGAU---- (((((((((.((..(((.(((..((((((((......)))))))).)))..)))...((((....(((((....-.)))))(((...)))...--.)))))).)))))))))..---- ( -41.20, z-score = -2.31, R) >droSim1.chr3R 18567263 111 - 27517382 CAUUUUGGAACCAAGCCCAUCUUCUGGCCACAGAUUUGUGGCCAGCGAUAUGGCCAAAUGGAACAUGUUCCCGC-CGAACAGAUGAUGUCAAU--GCCAUGGGGCUAAGAUGAA---- ((((((((..((..(((.(((..((((((((......)))))))).)))..)))...((((....(((((....-.)))))(((...)))...--.))))))..))))))))..---- ( -40.10, z-score = -2.42, R) >droVir3.scaffold_12822 3957468 90 + 4096053 CGUUUUGACAGGAUACCCAGAUUGUAGCU-UAUAAUAAGAACAUGAGGUUAGAAAAGACAGAUAAUACUAGUGCUCAAAGGUCUCACGUCA--------------------------- .....((((..((.(((...((((((...-))))))..((.(((.((((((...........)))).)).))).))...))).))..))))--------------------------- ( -11.40, z-score = 1.51, R) >consensus CAUUUUGGCACCAAGCCCAUCUUCUGGCCACAGAUUUGUGGCCAGCGAUAUGGCCAAAUGGAACAUGUUCCCGC_CGAACAGAUGAUGUCAAU__GCCAUGGGGCUA_G_UG______ ....((((((.((.(((......((((((((......))))))))......))).....((((....))))............)).)))))).......................... (-17.40 = -18.43 + 1.03)

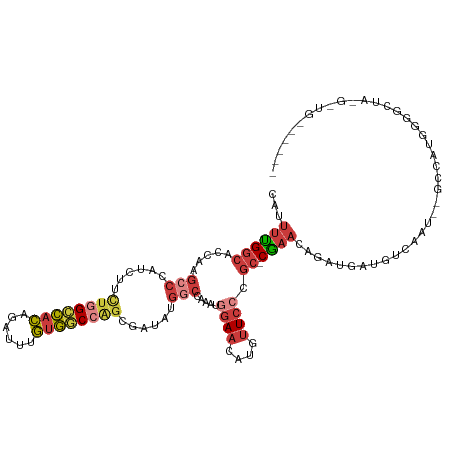
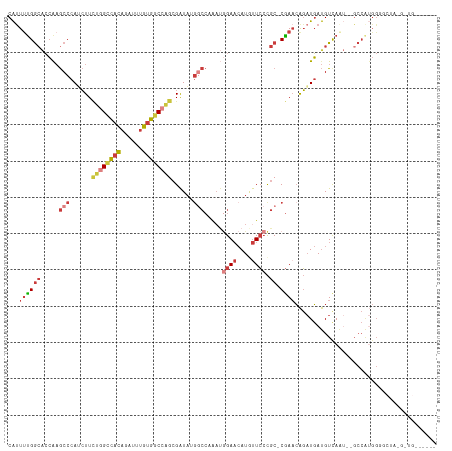
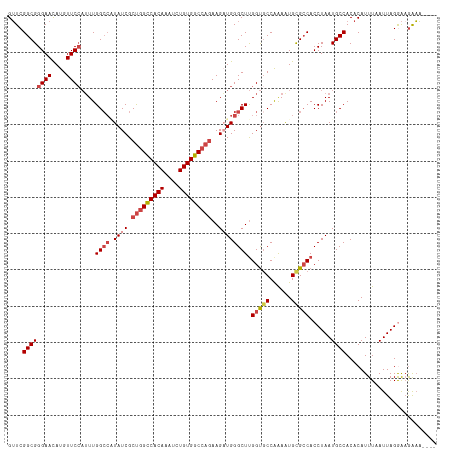
| Location | 18,756,232 – 18,756,346 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 89.54 |
| Shannon entropy | 0.19829 |
| G+C content | 0.49693 |
| Mean single sequence MFE | -43.02 |
| Consensus MFE | -33.71 |
| Energy contribution | -34.85 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
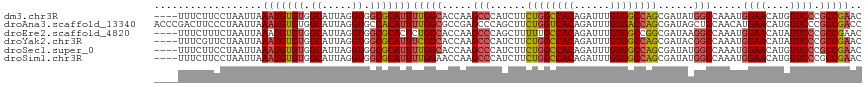
>dm3.chr3R 18756232 114 + 27905053 GUUCGGCGGGAACAUGUUCCAUUUGGCCAUAUCGCUGGCCACAAAUCUGUGGCCAGAAGAUGGGCUUGGUGCCAAAAUGCGCCACCUAAUGCCACACAUUUAAUUAGGAAGAAA---- .(((((((((((....))))((..((((.((((.(((((((((....)))))))))..))))))))..)).........)))).((((((............))))))..))).---- ( -44.70, z-score = -3.27, R) >droAna3.scaffold_13340 11131709 118 - 23697760 GGUCGGCGGGAACAUGUUCAUGUUGGAGCUAUCGCUGGUCACAAAUCUGUGACCAGAAGCUGGGCUCGGCGCCAAAAUGUGGCACCUAAUGCCACACAUUUAAUUAGGGAAGUCGGGU ..((((((.(((....))).))))))((((....(((((((((....))))))))).))))..(((((((((((.....)))).((((((............))))))...))))))) ( -45.00, z-score = -2.64, R) >droEre2.scaffold_4820 1157825 114 - 10470090 GUUCGGCGGGAAUAUGUUCCAUUUGGCCUUAUCGCCGGCCACAAAUCUGUGGCAAAAAGCUGGGCUUGGUGCCAGAGUGCGCCACCUAAUGCCACACAUUUAAUUAGAAAGAAA---- (((((((.((((....))))...((((......))))((((((....)))))).....))))))).((((((......)))))).(((((............))))).......---- ( -36.40, z-score = -0.89, R) >droYak2.chr3R 19613592 114 + 28832112 GUUCGGCGGGAAUAUGUUCCAUUUGGCCGUAUCGCUGGCCACAAAUCUGUGGCCAGAAGAUGGGCUUGGUGCCGAAAUGCGCCACCUAAUGCCACACAUUUAAUUAGAACGAAA---- ((((((((((((....))))....((((.((((.(((((((((....)))))))))..))))))))((((((......)))))).....))))....(......).))))....---- ( -43.20, z-score = -2.93, R) >droSec1.super_0 19121044 114 + 21120651 GUUCGGCGGGAACAUGUUCCAUUUGGCCAUAUCGCUGGCCACAAAUCUGUGGCCAGAAGAUGGGCUUGGUGCCAAAAUGCGCCACCUAAUGCCACACAUUUAAUUAGGAAGAAA---- .(((((((((((....))))((..((((.((((.(((((((((....)))))))))..))))))))..)).........)))).((((((............))))))..))).---- ( -44.70, z-score = -3.27, R) >droSim1.chr3R 18567295 114 + 27517382 GUUCGGCGGGAACAUGUUCCAUUUGGCCAUAUCGCUGGCCACAAAUCUGUGGCCAGAAGAUGGGCUUGGUUCCAAAAUGCGCCACCUAAUGCCACACAUUUAAUUAGGAAGAAA---- .(((((((((((....))))((..((((.((((.(((((((((....)))))))))..))))))))..)).........)))).((((((............))))))..))).---- ( -44.10, z-score = -3.49, R) >consensus GUUCGGCGGGAACAUGUUCCAUUUGGCCAUAUCGCUGGCCACAAAUCUGUGGCCAGAAGAUGGGCUUGGUGCCAAAAUGCGCCACCUAAUGCCACACAUUUAAUUAGGAAGAAA____ ....((((((((....))))....((((.((((.(((((((((....)))))))))..)))))))).(((((......)))))......))))......................... (-33.71 = -34.85 + 1.14)


| Location | 18,756,232 – 18,756,346 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 89.54 |
| Shannon entropy | 0.19829 |
| G+C content | 0.49693 |
| Mean single sequence MFE | -39.87 |
| Consensus MFE | -32.86 |
| Energy contribution | -33.37 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.977537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
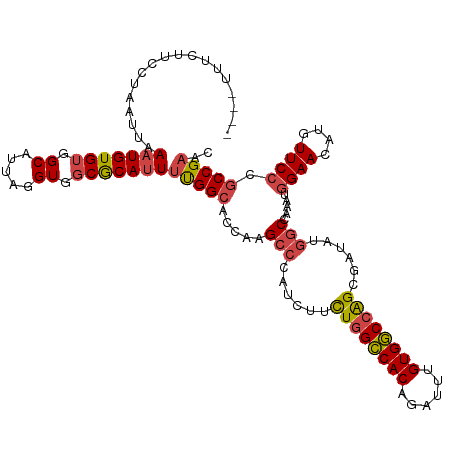
>dm3.chr3R 18756232 114 - 27905053 ----UUUCUUCCUAAUUAAAUGUGUGGCAUUAGGUGGCGCAUUUUGGCACCAAGCCCAUCUUCUGGCCACAGAUUUGUGGCCAGCGAUAUGGCCAAAUGGAACAUGUUCCCGCCGAAC ----..............(((((((.((.....)).)))))))(((((.....(((.(((..((((((((......)))))))).)))..))).....((((....)))).))))).. ( -40.60, z-score = -2.42, R) >droAna3.scaffold_13340 11131709 118 + 23697760 ACCCGACUUCCCUAAUUAAAUGUGUGGCAUUAGGUGCCACAUUUUGGCGCCGAGCCCAGCUUCUGGUCACAGAUUUGUGACCAGCGAUAGCUCCAACAUGAACAUGUUCCCGCCGACC .....................(((((((((...))))))))).((((((..((((.......((((((((......)))))))).....)))).(((((....)))))..)))))).. ( -39.60, z-score = -3.62, R) >droEre2.scaffold_4820 1157825 114 + 10470090 ----UUUCUUUCUAAUUAAAUGUGUGGCAUUAGGUGGCGCACUCUGGCACCAAGCCCAGCUUUUUGCCACAGAUUUGUGGCCGGCGAUAAGGCCAAAUGGAACAUAUUCCCGCCGAAC ----.....(((....(((((.(((((((..((.(((.((....((....)).))))).))...))))))).))))).(((.(((......)))....((((....)))).)))))). ( -36.40, z-score = -1.40, R) >droYak2.chr3R 19613592 114 - 28832112 ----UUUCGUUCUAAUUAAAUGUGUGGCAUUAGGUGGCGCAUUUCGGCACCAAGCCCAUCUUCUGGCCACAGAUUUGUGGCCAGCGAUACGGCCAAAUGGAACAUAUUCCCGCCGAAC ----....((((.....((((((((.((.....)).)))))))).(((.....(((.(((..((((((((......)))))))).)))..))).....((((....)))).))))))) ( -41.50, z-score = -3.00, R) >droSec1.super_0 19121044 114 - 21120651 ----UUUCUUCCUAAUUAAAUGUGUGGCAUUAGGUGGCGCAUUUUGGCACCAAGCCCAUCUUCUGGCCACAGAUUUGUGGCCAGCGAUAUGGCCAAAUGGAACAUGUUCCCGCCGAAC ----..............(((((((.((.....)).)))))))(((((.....(((.(((..((((((((......)))))))).)))..))).....((((....)))).))))).. ( -40.60, z-score = -2.42, R) >droSim1.chr3R 18567295 114 - 27517382 ----UUUCUUCCUAAUUAAAUGUGUGGCAUUAGGUGGCGCAUUUUGGAACCAAGCCCAUCUUCUGGCCACAGAUUUGUGGCCAGCGAUAUGGCCAAAUGGAACAUGUUCCCGCCGAAC ----....((((....(((((.((((((...((((((.((...(((....)))))))))))....)))))).)))))((((((......))))))...))))...((((.....)))) ( -40.50, z-score = -2.67, R) >consensus ____UUUCUUCCUAAUUAAAUGUGUGGCAUUAGGUGGCGCAUUUUGGCACCAAGCCCAUCUUCUGGCCACAGAUUUGUGGCCAGCGAUAUGGCCAAAUGGAACAUGUUCCCGCCGAAC ..................(((((((.((.....)).)))))))(((((.....(((......((((((((......))))))))......))).....((((....)))).))))).. (-32.86 = -33.37 + 0.50)
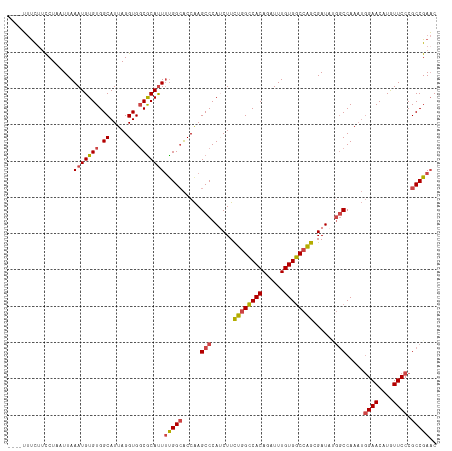
| Location | 18,756,271 – 18,756,384 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 80.86 |
| Shannon entropy | 0.36201 |
| G+C content | 0.44852 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -21.28 |
| Energy contribution | -22.03 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.940355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18756271 113 - 27905053 AAUAUUGUGGUUUGGGUUC-UUUCUCCUCAAUUUCCCAGUUUC----UUCCUAAUUAAAUGUGUGGCAUUAGGUGGCGCAUUUUGGCACCAAGCCCAUCUUCUGGCCACAGAUUUGUG ...((((.((.(((((...-.....)).)))...))))))...----........(((((.((((((...((((((.((...(((....)))))))))))....)))))).))))).. ( -32.40, z-score = -2.16, R) >droAna3.scaffold_13340 11131748 118 + 23697760 AAAAUGGUUGGCUUUCGAUUCUUUUUUUCGCUUUCCAUGACCCGACUUCCCUAAUUAAAUGUGUGGCAUUAGGUGCCACAUUUUGGCGCCGAGCCCAGCUUCUGGUCACAGAUUUGUG ...((((..(((.................)))..)))).................(((((.((((((....(((((((.....)))))))((((...))))...)))))).))))).. ( -32.13, z-score = -1.44, R) >droEre2.scaffold_4820 1157864 114 + 10470090 AAAAUCGUGGGUUGGAUUUGCUUCUCCCCAAUUUCCCCGUUUC----UUUCUAAUUAAAUGUGUGGCAUUAGGUGGCGCACUCUGGCACCAAGCCCAGCUUUUUGCCACAGAUUUGUG .......((((..(((.....)))..)))).............----........(((((.(((((((..((.(((.((....((....)).))))).))...))))))).))))).. ( -30.90, z-score = -1.03, R) >droYak2.chr3R 19613631 102 - 28832112 -----------CUUUUCAU-UUUCUUCCCAAUUUCCCAGUUUC----GUUCUAAUUAAAUGUGUGGCAUUAGGUGGCGCAUUUCGGCACCAAGCCCAUCUUCUGGCCACAGAUUUGUG -----------........-.......................----........(((((.((((((...((((((.((.............))))))))....)))))).))))).. ( -24.12, z-score = -1.94, R) >droSec1.super_0 19121083 113 - 21120651 AAAAUUGUGGCUUGGGUUC-UUUCUACCCGAUUUCCCAGUUUC----UUCCUAAUUAAAUGUGUGGCAUUAGGUGGCGCAUUUUGGCACCAAGCCCAUCUUCUGGCCACAGAUUUGUG .((((((.((.((((((..-.....))))))...)))))))).----........(((((.((((((...((((((.((...(((....)))))))))))....)))))).))))).. ( -37.60, z-score = -3.09, R) >droSim1.chr3R 18567334 113 - 27517382 AAAAUUGUGGUUUGGGUUC-UUUCUCCCCGAUUUCCCAGUUUC----UUCCUAAUUAAAUGUGUGGCAUUAGGUGGCGCAUUUUGGAACCAAGCCCAUCUUCUGGCCACAGAUUUGUG .((((((.((.(((((...-......)))))...)))))))).----........(((((.((((((...((((((.((...(((....)))))))))))....)))))).))))).. ( -36.50, z-score = -3.02, R) >consensus AAAAUUGUGGGUUGGGUUC_UUUCUCCCCAAUUUCCCAGUUUC____UUCCUAAUUAAAUGUGUGGCAUUAGGUGGCGCAUUUUGGCACCAAGCCCAUCUUCUGGCCACAGAUUUGUG .......................................................(((((.((((((...((((((.((...(((....)))))))))))....)))))).))))).. (-21.28 = -22.03 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:36:40 2011