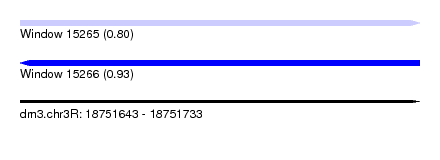
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,751,643 – 18,751,733 |
| Length | 90 |
| Max. P | 0.933675 |

| Location | 18,751,643 – 18,751,733 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 67.51 |
| Shannon entropy | 0.62373 |
| G+C content | 0.48764 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -12.74 |
| Energy contribution | -14.61 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.801075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
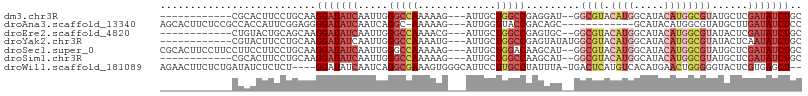
>dm3.chr3R 18751643 90 + 27905053 ------------CGCACUUCCUGCAAGGAUAUCAAUUGGGCCAAAAAG---AUUGCUGGCCGAGGAU--GGCGUACAUGGCAUACAUGGCGUAUGCUCGAUAUCUGC ------------.(((.....)))..(((((((.....(((((.....---.....)))))(((.((--(.(((.((((.....)))))))))).)))))))))).. ( -31.20, z-score = -1.86, R) >droAna3.scaffold_13340 11127199 91 - 23697760 AGCACUUCUCCGCCACCAUUCGGAGGGGAUAUCAAUCAGGC-AAAAAG---AUUGGUUACCGACAGC------------GCAUACAUGGCGUAUGCUUGAUAUCUCC ....((((((((........))))))))......(((((((-(.....---.((((...))))..((------------((.......)))).))))))))...... ( -26.00, z-score = -1.52, R) >droEre2.scaffold_4820 1153355 90 - 10470090 ------------CUGUACUGCAGCAAGGAUAUCAAUUGGGCCAAAACG---AUUGCUGGCCGAGUGC--GGCGUACAUGGCAUACAUGGCGUAUACUCGAUAUCUGC ------------(((.....)))...(((((((.....(((((.....---.....)))))(((((.--.((((.((((.....)))))))).)))))))))))).. ( -31.10, z-score = -1.80, R) >droYak2.chr3R 19608934 92 + 28832112 ------------CGUACUUCCUGCAAGGAUAUCAAUUGGGCCAAAAUG---AUUGCUGGCCGAGUAUAUGGCGUACAUGGCAUACAUGGCGUAUACUCAAUAUCUGC ------------.(((.....)))..((((((......(((((.....---.....)))))(((((((((.....((((.....)))).))))))))).)))))).. ( -26.10, z-score = -0.87, R) >droSec1.super_0 19116448 102 + 21120651 CGCACUUCCUUCCUUCCUUCCUGCAAGGAUAUCAAUUGGGCCAAAAAG---AUUGCUGGACAAGCAU--GGCGUACAUGGCAUACAUGGCGUAUGCUCGAUAUCUGC .(((.................)))..(((((((...((.((((.....---..((((.....)))))--))).))...(((((((.....))))))).))))))).. ( -25.64, z-score = 0.21, R) >droSim1.chr3R 18562727 90 + 27517382 ------------CGCACUUCCUGCAAGGAUAUCAAUUGGGCCAAAAAG---AUUGCUGGCCAAGCAU--GGCGUACAUGGCAUACAUGGCGUAUGCUCGAUAUCUGC ------------.(((.....)))..(((((((.....(((((.....---.....))))).(((((--(.(((.((((.....))))))))))))).))))))).. ( -31.20, z-score = -1.83, R) >droWil1.scaffold_181089 11087977 100 - 12369635 AGAACUUCUCUGAUAUCUCUCU----GGAUAUCAAUCAGGCGAAAGUGGGCAUUCCUUGCCUAUUUA-UGACUCAUGUCACAUGAACUGGGGGUACUCGUGAGCU-- ((.(((((..((((((((....----))))))))..(((....(((((((((.....))))))))).-((((....))))......)))))))).))(....)..-- ( -27.90, z-score = -1.14, R) >consensus ____________CGAACUUCCUGCAAGGAUAUCAAUUGGGCCAAAAAG___AUUGCUGGCCGAGUAU__GGCGUACAUGGCAUACAUGGCGUAUGCUCGAUAUCUGC ..........................(((((((.....(((((.............))))).........((((.((((.....))))))))......))))))).. (-12.74 = -14.61 + 1.86)


| Location | 18,751,643 – 18,751,733 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 67.51 |
| Shannon entropy | 0.62373 |
| G+C content | 0.48764 |
| Mean single sequence MFE | -25.70 |
| Consensus MFE | -15.48 |
| Energy contribution | -16.33 |
| Covariance contribution | 0.85 |
| Combinations/Pair | 1.26 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
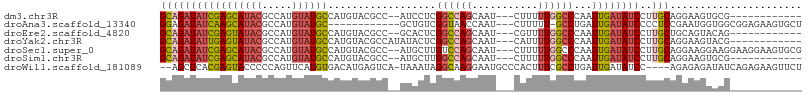
>dm3.chr3R 18751643 90 - 27905053 GCAGAUAUCGAGCAUACGCCAUGUAUGCCAUGUACGCC--AUCCUCGGCCAGCAAU---CUUUUUGGCCCAAUUGAUAUCCUUGCAGGAAGUGCG------------ (((((((((((((((((.....))))))..........--......((((((....---....))))))...))))))))..)))..........------------ ( -25.90, z-score = -1.19, R) >droAna3.scaffold_13340 11127199 91 + 23697760 GGAGAUAUCAAGCAUACGCCAUGUAUGC------------GCUGUCGGUAACCAAU---CUUUUU-GCCUGAUUGAUAUCCCCUCCGAAUGGUGGCGGAGAAGUGCU ((.((((((((((((((.....))))))------------......(((((.....---....))-)))...))))))))))(((((........)))))....... ( -28.70, z-score = -1.63, R) >droEre2.scaffold_4820 1153355 90 + 10470090 GCAGAUAUCGAGUAUACGCCAUGUAUGCCAUGUACGCC--GCACUCGGCCAGCAAU---CGUUUUGGCCCAAUUGAUAUCCUUGCUGCAGUACAG------------ (((((((((((((....((..(((((.....)))))..--))))))((((((....---....)))))).....))))))..)))..........------------ ( -24.60, z-score = -0.83, R) >droYak2.chr3R 19608934 92 - 28832112 GCAGAUAUUGAGUAUACGCCAUGUAUGCCAUGUACGCCAUAUACUCGGCCAGCAAU---CAUUUUGGCCCAAUUGAUAUCCUUGCAGGAAGUACG------------ ((((((((((((((((.((((((.....))))...))..)))))))((((((....---....)))))).....))))))..)))..........------------ ( -25.30, z-score = -1.59, R) >droSec1.super_0 19116448 102 - 21120651 GCAGAUAUCGAGCAUACGCCAUGUAUGCCAUGUACGCC--AUGCUUGUCCAGCAAU---CUUUUUGGCCCAAUUGAUAUCCUUGCAGGAAGGAAGGAAGGAAGUGCG (((..((((((((((((.....)))))).......(((--(((((.....))))..---.....))))....))))))(((((.(.....).)))))......))). ( -26.21, z-score = 0.32, R) >droSim1.chr3R 18562727 90 - 27517382 GCAGAUAUCGAGCAUACGCCAUGUAUGCCAUGUACGCC--AUGCUUGGCCAGCAAU---CUUUUUGGCCCAAUUGAUAUCCUUGCAGGAAGUGCG------------ (((((((((((((((((.....))))))((((.....)--)))...((((((....---....))))))...))))))))..)))..........------------ ( -26.60, z-score = -0.78, R) >droWil1.scaffold_181089 11087977 100 + 12369635 --AGCUCACGAGUACCCCCAGUUCAUGUGACAUGAGUCA-UAAAUAGGCAAGGAAUGCCCACUUUCGCCUGAUUGAUAUCC----AGAGAGAUAUCAGAGAAGUUCU --..(((..................((((((....))))-))..(((((((((........)))).)))))..(((((((.----.....))))))))))....... ( -22.60, z-score = -0.98, R) >consensus GCAGAUAUCGAGCAUACGCCAUGUAUGCCAUGUACGCC__AUACUCGGCCAGCAAU___CUUUUUGGCCCAAUUGAUAUCCUUGCAGGAAGUAAG____________ (((((((((((((((((.....))))))..................((((((...........))))))...))))))))..)))...................... (-15.48 = -16.33 + 0.85)
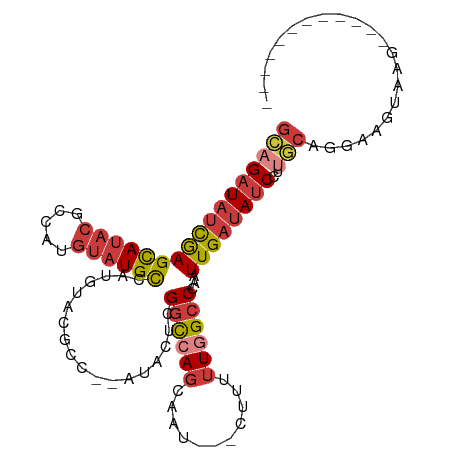
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:36:35 2011