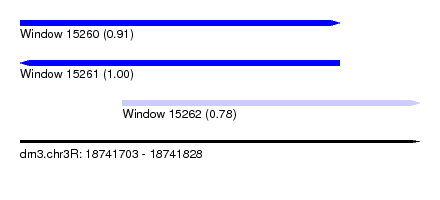
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,741,703 – 18,741,828 |
| Length | 125 |
| Max. P | 0.997827 |

| Location | 18,741,703 – 18,741,803 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 88.46 |
| Shannon entropy | 0.21559 |
| G+C content | 0.45245 |
| Mean single sequence MFE | -29.74 |
| Consensus MFE | -21.66 |
| Energy contribution | -22.80 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.913756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
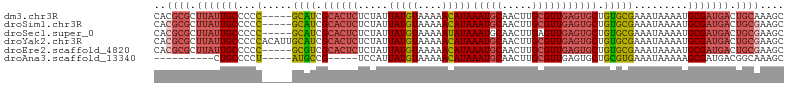
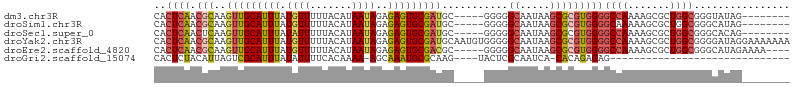
>dm3.chr3R 18741703 100 + 27905053 GCUUUGCAGUCAUCGCAUUUUAUUUCGCACAGCACUCAACGCAAGUUGCAUUUAUGUUUUUACAUAAUAGAGAGUGCGAUGC-----GGGGGCAAUAAGCGCGUG (((((((.((....))......(..((((..(((((((((....)))....((((((....))))))....))))))..)))-----)..))))..))))..... ( -32.80, z-score = -2.80, R) >droSim1.chr3R 18552828 100 + 27517382 GCUUCGCAGUCAUCGCAUUUUAUUUCGCACAGCACUCAACGCAAGUUGCAUUUAUGUUUUUACAUAAUAGAGAGUGCGAUGC-----GGGGGCAAUAAGCGCGUG .............(((......(..((((..(((((((((....)))....((((((....))))))....))))))..)))-----)..)((.....))))).. ( -31.90, z-score = -2.59, R) >droSec1.super_0 19106664 100 + 21120651 GCUUCGCAGUCAUCGCAUUUUAUUUCGCACAGCACUCAACUCAAGUUGCAUUUAUAUUUUUACAUAAUAGAGAGUGCGAUGC-----GGGGGCAAUAAGCGCGUG .............(((......(..((((..(((((((((....))).......((((.......))))..))))))..)))-----)..)((.....))))).. ( -24.10, z-score = -0.84, R) >droYak2.chr3R 19598670 105 + 28832112 GCUUCGCAGUCAUCGCAUUUUAUUUCGCACAGCACUCAACGCAAGUUGCAUUUAUGUUUUUACAUAAUAGAGAGUGCGAUGCAAUGUGGGGGCAAUAAGCGCGUG ((((....(((..((((((.......(((..(((((((((....)))....((((((....))))))....))))))..)))))))))..)))...))))..... ( -32.71, z-score = -2.14, R) >droEre2.scaffold_4820 1143552 100 - 10470090 GCUUCGCAGUCAUCGCAUUUUAUUUCGCACAGCACUCAACGCAAGUUGCAUUUAUGUUUUUACAUAAUAGAGAGUGCGACGC-----GGGGGCAAUAAGCGCGUG .((((((.(((...((..........))...(((((((((....)))....((((((....))))))....)))))))))))-----))))((.....))..... ( -31.10, z-score = -2.43, R) >droAna3.scaffold_13340 11117337 85 - 23697760 GCUUUGCCGUCAUCGCUUUUUAUUUCACGCAGCACUCAACGCAAGUUGCAUUUAUGUUUUUACAUAAUGGA-----CGGCAU-----AGGGGCAG---------- (((((((((((...(((.............)))...((((....))))((((.((((....))))))))))-----))))).-----..))))..---------- ( -25.82, z-score = -3.65, R) >consensus GCUUCGCAGUCAUCGCAUUUUAUUUCGCACAGCACUCAACGCAAGUUGCAUUUAUGUUUUUACAUAAUAGAGAGUGCGAUGC_____GGGGGCAAUAAGCGCGUG (((((.....(((((((((((...............((((....))))...((((((....))))))...))))))))))).......)))))............ (-21.66 = -22.80 + 1.14)

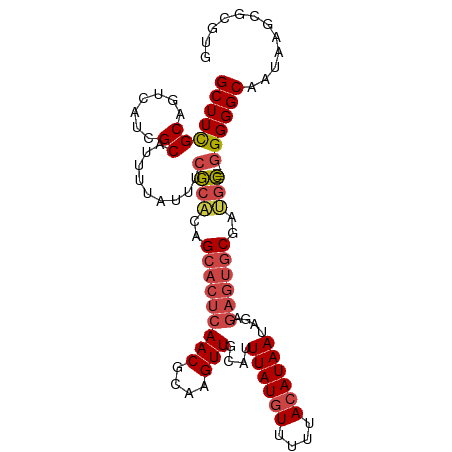
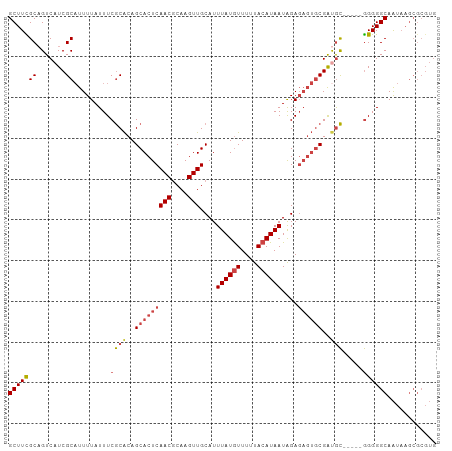
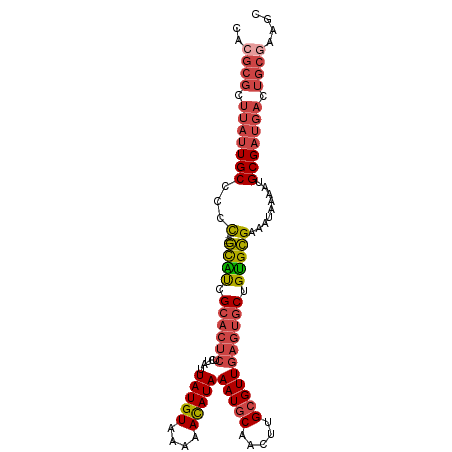
| Location | 18,741,703 – 18,741,803 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 88.46 |
| Shannon entropy | 0.21559 |
| G+C content | 0.45245 |
| Mean single sequence MFE | -30.68 |
| Consensus MFE | -24.88 |
| Energy contribution | -26.33 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.61 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.19 |
| SVM RNA-class probability | 0.997827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18741703 100 - 27905053 CACGCGCUUAUUGCCCCC-----GCAUCGCACUCUCUAUUAUGUAAAAACAUAAAUGCAACUUGCGUUGAGUGCUGUGCGAAAUAAAAUGCGAUGACUGCAAAGC ...(((.(((((((...(-----((((.((((((.....(((((....)))))(((((.....))))))))))).))))).........))))))).)))..... ( -31.80, z-score = -3.86, R) >droSim1.chr3R 18552828 100 - 27517382 CACGCGCUUAUUGCCCCC-----GCAUCGCACUCUCUAUUAUGUAAAAACAUAAAUGCAACUUGCGUUGAGUGCUGUGCGAAAUAAAAUGCGAUGACUGCGAAGC ..((((.(((((((...(-----((((.((((((.....(((((....)))))(((((.....))))))))))).))))).........))))))).)))).... ( -33.80, z-score = -4.41, R) >droSec1.super_0 19106664 100 - 21120651 CACGCGCUUAUUGCCCCC-----GCAUCGCACUCUCUAUUAUGUAAAAAUAUAAAUGCAACUUGAGUUGAGUGCUGUGCGAAAUAAAAUGCGAUGACUGCGAAGC ..((((.(((((((...(-----((((.((((((....((((((....))))))....(((....))))))))).))))).........))))))).)))).... ( -28.80, z-score = -3.14, R) >droYak2.chr3R 19598670 105 - 28832112 CACGCGCUUAUUGCCCCCACAUUGCAUCGCACUCUCUAUUAUGUAAAAACAUAAAUGCAACUUGCGUUGAGUGCUGUGCGAAAUAAAAUGCGAUGACUGCGAAGC ..((((.(((((((.......((((((.((((((.....(((((....)))))(((((.....))))))))))).))))))........))))))).)))).... ( -35.66, z-score = -4.65, R) >droEre2.scaffold_4820 1143552 100 + 10470090 CACGCGCUUAUUGCCCCC-----GCGUCGCACUCUCUAUUAUGUAAAAACAUAAAUGCAACUUGCGUUGAGUGCUGUGCGAAAUAAAAUGCGAUGACUGCGAAGC ..((((.(((((((...(-----(((..((((((.....(((((....)))))(((((.....)))))))))))..)))).........))))))).)))).... ( -32.10, z-score = -3.46, R) >droAna3.scaffold_13340 11117337 85 + 23697760 ----------CUGCCCCU-----AUGCCG-----UCCAUUAUGUAAAAACAUAAAUGCAACUUGCGUUGAGUGCUGCGUGAAAUAAAAAGCGAUGACGGCAAAGC ----------......((-----.(((((-----((..((((((....))))))(((((((((.....))))..)))))...............))))))).)). ( -21.90, z-score = -2.13, R) >consensus CACGCGCUUAUUGCCCCC_____GCAUCGCACUCUCUAUUAUGUAAAAACAUAAAUGCAACUUGCGUUGAGUGCUGUGCGAAAUAAAAUGCGAUGACUGCGAAGC ..((((.(((((((.........((((.((((((.....(((((....)))))(((((.....))))))))))).))))..........))))))).)))).... (-24.88 = -26.33 + 1.45)
| Location | 18,741,735 – 18,741,828 |
|---|---|
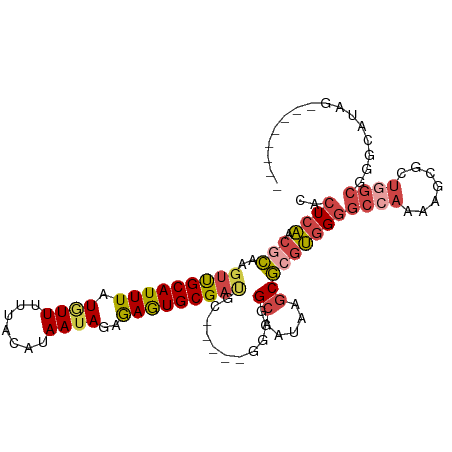
| Length | 93 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 76.22 |
| Shannon entropy | 0.43201 |
| G+C content | 0.47978 |
| Mean single sequence MFE | -25.57 |
| Consensus MFE | -15.41 |
| Energy contribution | -17.11 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.780814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18741735 93 + 27905053 CACUCAACGCAAGUUGCAUUUAUGUUUUUACAUAAUAGAGAGUGCGAUGC-----GGGGGCAAUAAGCGCGUGGGGCCAAAAGCGCUGGCGGGUAUAG-------- .((((..((((..((((((((.((((.......))))..)))))))))))-----)...((....(((((.((....))...))))).))))))....-------- ( -29.60, z-score = -1.83, R) >droSim1.chr3R 18552860 93 + 27517382 CACUCAACGCAAGUUGCAUUUAUGUUUUUACAUAAUAGAGAGUGCGAUGC-----GGGGGCAAUAAGCGCGUGGGGCAAAAAGCGCUGGCGGGCAUAG-------- ((((((((....)))....((((((....))))))....)))))...(((-----....)))....((.(((.((((.....)).)).))).))....-------- ( -28.10, z-score = -1.65, R) >droSec1.super_0 19106696 93 + 21120651 CACUCAACUCAAGUUGCAUUUAUAUUUUUACAUAAUAGAGAGUGCGAUGC-----GGGGGCAAUAAGCGCGUGGGGCCAAAAGCGCUGGCGGGCACAG-------- ..(((..(((..(((((((((.((((.......))))..)))))))))..-----.)))((....(((((.((....))...))))).))))).....-------- ( -27.90, z-score = -1.58, R) >droYak2.chr3R 19598702 106 + 28832112 CACUCAACGCAAGUUGCAUUUAUGUUUUUACAUAAUAGAGAGUGCGAUGCAAUGUGGGGGCAAUAAGCGCGUGGGGCCAAAAGCGCUGGCGGGGAUAGGAAAAAAA ..(((..((((.(((((((((.((((.......))))..)))))))))....))))...((....(((((.((....))...))))).)))))............. ( -28.40, z-score = -1.06, R) >droEre2.scaffold_4820 1143584 97 - 10470090 CACUCAACGCAAGUUGCAUUUAUGUUUUUACAUAAUAGAGAGUGCGACGC-----GGGGGCAAUAAGCGCGUGGGGCCAAAAGCGCUGGCGGGCAUAGAAAA---- ..(((..(((..(((((((((.((((.......))))..)))))))))))-----)...((....(((((.((....))...))))).))))).........---- ( -28.90, z-score = -1.16, R) >droGri2.scaffold_15074 3546118 70 + 7742996 CACUCUACAUUAGUCGCAUUUAUAUUUUCACAAAA-AGCAAAUGCGCAAG----UACUCGCAAUCA-CACAGAGAG------------------------------ ..((((.........((((((...((((....)))-)..))))))((...----.....)).....-...))))..------------------------------ ( -10.50, z-score = -1.96, R) >consensus CACUCAACGCAAGUUGCAUUUAUGUUUUUACAUAAUAGAGAGUGCGAUGC_____GGGGGCAAUAAGCGCGUGGGGCCAAAAGCGCUGGCGGGCAUAG________ ..(((.......(((((((((.((((.......))))..)))))))))........))).......((.(((.((((.....)).)).))).))............ (-15.41 = -17.11 + 1.70)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:36:32 2011