
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,737,053 – 18,737,158 |
| Length | 105 |
| Max. P | 0.941075 |

| Location | 18,737,053 – 18,737,158 |
|---|---|
| Length | 105 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 71.71 |
| Shannon entropy | 0.61148 |
| G+C content | 0.48867 |
| Mean single sequence MFE | -31.83 |
| Consensus MFE | -15.21 |
| Energy contribution | -15.61 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.941075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
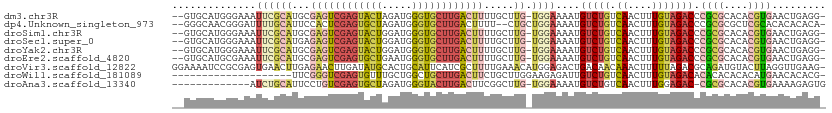
>dm3.chr3R 18737053 105 + 27905053 --GUGCAUGGGAAAUUCGCAUGCGAGUCGAGUACUAGAUGGGUGCUUGACUUUUGCUUG-UGGAAAAUGUCUGUCAACUUUGUAGACCCGCGCACACGUGAACUGAGG- --((((.((((...((((((.((((((((((((((.....))))))))))))..)).))-)))).....((((.((....)))))))))).)))).............- ( -38.50, z-score = -3.02, R) >dp4.Unknown_singleton_973 8343 104 - 14580 --GGGCAACGGGAUUUUGCAUUCCACUCGAGUGCUAGAUGGGUGCUUGACUUUU--CUGCUGGAAAAUGUCUGUCAACUUUGUAGACCCGCGCGCUCGCACACACACA- --((((..((((((((.((((((.....)))))).))))((((....(((((((--(.....))))).))).....))))......))))...))))...........- ( -27.10, z-score = 0.55, R) >droSim1.chr3R 18548202 105 + 27517382 --GUGCAUGGGAAAUUCGCAUGCGAGUCGAGUACUGGAUGGGUGCUUGACUUUUGCUUG-UGGAAAAUGUCUGUCAACUUUGUAGACCCGCGCACACGUGAACUGAGG- --((((.((((...((((((.((((((((((((((.....))))))))))))..)).))-)))).....((((.((....)))))))))).)))).............- ( -38.20, z-score = -2.76, R) >droSec1.super_0 19102052 105 + 21120651 --GUGCAUGGGAAAUUCGCAUGAGAGUCGAGUACUGGAUGGGUGCUUGACUUUUGCUUG-UGGAAAAUGUCUGUCAACUUUGUAGACCCGCGCACACGUGAACUGAGG- --((((.((((...((((((.((((((((((((((.....))))))))))))))...))-)))).....((((.((....)))))))))).)))).............- ( -37.70, z-score = -2.89, R) >droYak2.chr3R 19593810 105 + 28832112 --GUGCAUGGGAAAUUCGCAUGCGAGUCGAGUACUGGAUGGGUGCUUGACUUUUGCUUG-UGGAAAAUGUCUGUCAACUUUGUAGACCCGCGCACACGUGAACUGAGG- --((((.((((...((((((.((((((((((((((.....))))))))))))..)).))-)))).....((((.((....)))))))))).)))).............- ( -38.20, z-score = -2.76, R) >droEre2.scaffold_4820 1138939 105 - 10470090 --GUGCAUGCGAAAUUCGCAUGCGAGUCGAGUGCUGAAUGGGUGCUUGACUUUUGCUUG-UGGAAAAUGUCUGUCAACUUUGUAGACCCGCGCACACGUGAACUGAGG- --..(((((((.....)))))))((((((((..((.....))..))))))))(..(.((-(((.....(((((.((....)))))))...).)))).)..).......- ( -37.30, z-score = -2.33, R) >droVir3.scaffold_12822 3942743 108 - 4096053 GGAAAAUCCGCGAGUGAACUUGAGAACUUGAUAUGCACUGCAUUCAUCGCUUUUGAAACAUGGAGACUGACAACAAACUUUUUAGACGCAGAUGUACUUAGGUUGAAG- ((.....)).((((....))))..(((((((..((((((((.((((.......)))).((.(....)))..................)))).)))).)))))))....- ( -22.10, z-score = -0.64, R) >droWil1.scaffold_181089 11072605 88 - 12369635 --------------------UUCGGGUCGAGUGUUUGCUGGCUGCUUGACUUCUGCUUGGAAGAGAUUGUCUGUCAACUUUGUAGACACACACACACAUGAACACACG- --------------------.......((.(((((((.((..((..((.(((((....)))))....((((((.((....))))))))))..))..))))))))).))- ( -18.70, z-score = 1.33, R) >droAna3.scaffold_13340 11112821 94 - 23697760 -------------AUCUGCAUUCCUGUCGAGUGCUAGAUGGGUACUUGACUUCGGCUUG-UGGAAAAUGUCUGUCAACUUUGGAGAC-CGCGCACACGUGAAAAGAGUG -------------.((..((..((.((((((((((.....))))))))))...))..))-..))....((((.(((....)))))))-((((....))))......... ( -28.70, z-score = -1.32, R) >consensus __GUGCAUGGGAAAUUCGCAUGCGAGUCGAGUACUGGAUGGGUGCUUGACUUUUGCUUG_UGGAAAAUGUCUGUCAACUUUGUAGACCCGCGCACACGUGAACUGAGG_ .......................((((((((((((.....))))))))))))................(((((.((....))))))).((((....))))......... (-15.21 = -15.61 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:36:30 2011