| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,717,470 – 18,717,544 |
| Length | 74 |
| Max. P | 0.998307 |

| Location | 18,717,470 – 18,717,544 |
|---|---|
| Length | 74 |
| Sequences | 6 |
| Columns | 81 |
| Reading direction | forward |
| Mean pairwise identity | 71.56 |
| Shannon entropy | 0.51858 |
| G+C content | 0.50720 |
| Mean single sequence MFE | -23.08 |
| Consensus MFE | -16.61 |
| Energy contribution | -19.78 |
| Covariance contribution | 3.17 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.32 |
| SVM RNA-class probability | 0.998307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
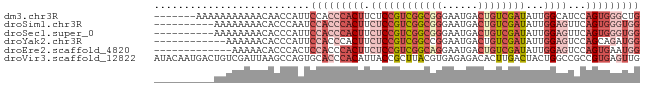
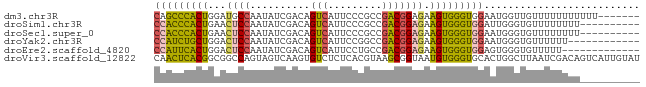
>dm3.chr3R 18717470 74 + 27905053 -------AAAAAAAAAAACAACCAUUCCACCCACUUCUCCGUCGGCGGGAAUGACUGUCGAUAUUGGCAUCCAGUGGGCUG -------......................((((((...((((((((((......))))))))...)).....))))))... ( -20.20, z-score = -1.75, R) >droSim1.chr3R 18528733 71 + 27517382 ----------AAAAAAAACACCCAAUCCACCCACUUCUCCGUCGGCGGGAAUGACUGUCGAUAUUGGAGUUCAGUGGGUGG ----------................(((((((((.((((((((((((......))))))))...))))...))))))))) ( -32.80, z-score = -5.08, R) >droSec1.super_0 19082719 71 + 21120651 ----------AAAAAAAACACCCAUUCCACCCACUUCUCCGUCGGCGGGAAUGACUGUCGAUAUUGGAGUUCAGUGGGUGG ----------................(((((((((.((((((((((((......))))))))...))))...))))))))) ( -32.80, z-score = -4.95, R) >droYak2.chr3R 19571630 69 + 28832112 ------------AAAAAACACCCAUUCCACCCACUUCUCCGUCGGCCGGAAUGACUGUCGAUAUUGGAGUCCAGCAGAUGG ------------.........(((((......(((((...((((((.(......).))))))...)))))......))))) ( -15.90, z-score = -0.50, R) >droEre2.scaffold_4820 1119591 68 - 10470090 -------------AAAAACACCCACUCCACCCACUUCUCCGUCGGCAGGAAUGACUGUCGAUAUUGGAGUCCAGUGAAUGG -------------.............(((..((((.((((((((((((......))))))))...))))...))))..))) ( -23.10, z-score = -2.91, R) >droVir3.scaffold_12822 3928804 81 - 4096053 AUACAAUGACUGUCGAUUAAGCCAGUGCACCCACAUUACCGCUUACGUGAGAGACACUUGACUACUGGCCGCCGUGAGUUG ...(((((.((.(((..(((((.((((......))))...)))))..))).)).)).)))...(((.((....)).))).. ( -13.70, z-score = 0.75, R) >consensus __________AAAAAAAACACCCAUUCCACCCACUUCUCCGUCGGCGGGAAUGACUGUCGAUAUUGGAGUCCAGUGGGUGG ..........................(((((((((.((((((((((((......))))))))...))))...))))))))) (-16.61 = -19.78 + 3.17)
| Location | 18,717,470 – 18,717,544 |
|---|---|
| Length | 74 |
| Sequences | 6 |
| Columns | 81 |
| Reading direction | reverse |
| Mean pairwise identity | 71.56 |
| Shannon entropy | 0.51858 |
| G+C content | 0.50720 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -14.53 |
| Energy contribution | -14.48 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.835258 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
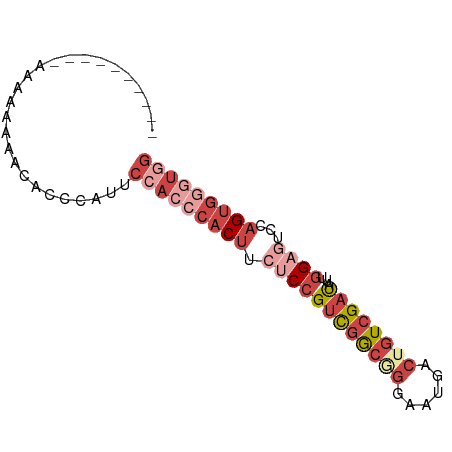
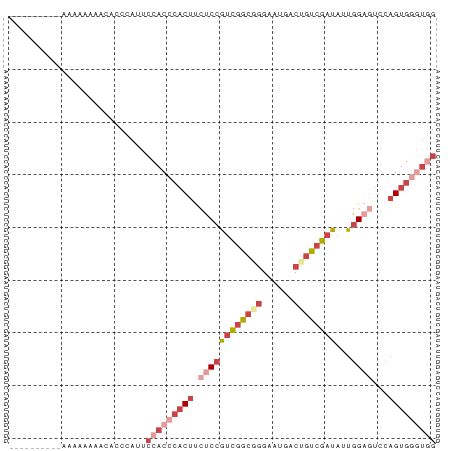
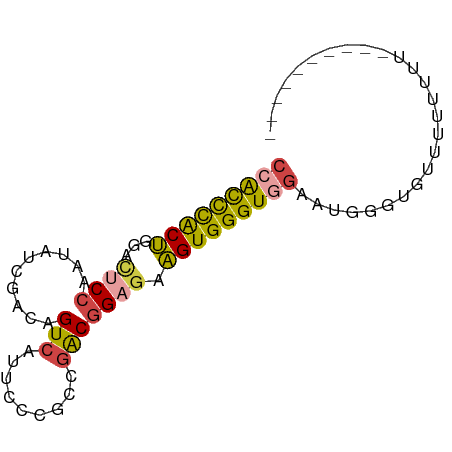
>dm3.chr3R 18717470 74 - 27905053 CAGCCCACUGGAUGCCAAUAUCGACAGUCAUUCCCGCCGACGGAGAAGUGGGUGGAAUGGUUGUUUUUUUUUUU------- (((....)))((((....))))((((..((((((.(((.((......)).)))))))))..)))).........------- ( -22.50, z-score = -1.43, R) >droSim1.chr3R 18528733 71 - 27517382 CCACCCACUGAACUCCAAUAUCGACAGUCAUUCCCGCCGACGGAGAAGUGGGUGGAUUGGGUGUUUUUUUU---------- (((((((((...((((....(((.(.(......).).))).)))).)))))))))................---------- ( -22.80, z-score = -1.40, R) >droSec1.super_0 19082719 71 - 21120651 CCACCCACUGAACUCCAAUAUCGACAGUCAUUCCCGCCGACGGAGAAGUGGGUGGAAUGGGUGUUUUUUUU---------- (((((((((...((((....(((.(.(......).).))).)))).)))))))))................---------- ( -22.80, z-score = -1.37, R) >droYak2.chr3R 19571630 69 - 28832112 CCAUCUGCUGGACUCCAAUAUCGACAGUCAUUCCGGCCGACGGAGAAGUGGGUGGAAUGGGUGUUUUUU------------ (((.....)))...........((((.((((((((.((.((......)).)))))))))).))))....------------ ( -21.80, z-score = -0.75, R) >droEre2.scaffold_4820 1119591 68 + 10470090 CCAUUCACUGGACUCCAAUAUCGACAGUCAUUCCUGCCGACGGAGAAGUGGGUGGAGUGGGUGUUUUU------------- (((((((((...((((....(((.(((......))).))).)))).))))))))).............------------- ( -22.00, z-score = -0.98, R) >droVir3.scaffold_12822 3928804 81 + 4096053 CAACUCACGGCGGCCAGUAGUCAAGUGUCUCUCACGUAAGCGGUAAUGUGGGUGCACUGGCUUAAUCGACAGUCAUUGUAU ..(((((((...(((.((......(((.....)))....)))))..)))))))(((.(((((........))))).))).. ( -20.50, z-score = -0.09, R) >consensus CCACCCACUGGACUCCAAUAUCGACAGUCAUUCCCGCCGACGGAGAAGUGGGUGGAAUGGGUGUUUUUUUU__________ (((((((((...((((..........(((.........))))))).))))))))).......................... (-14.53 = -14.48 + -0.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:36:27 2011