
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,705,284 – 18,705,357 |
| Length | 73 |
| Max. P | 0.696100 |

| Location | 18,705,284 – 18,705,357 |
|---|---|
| Length | 73 |
| Sequences | 8 |
| Columns | 78 |
| Reading direction | reverse |
| Mean pairwise identity | 80.66 |
| Shannon entropy | 0.38544 |
| G+C content | 0.39523 |
| Mean single sequence MFE | -18.79 |
| Consensus MFE | -11.70 |
| Energy contribution | -11.44 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.696100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
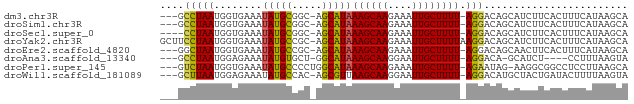
>dm3.chr3R 18705284 73 - 27905053 ---GCCUAAUGGUGAAAUAUGCGGC-AGCAUAAAGCAAGAAAUUGCUUUU-AGGACAGCAUCUUCACUUUCAUAAGCA ---((.((..((((((..((((..(-.....(((((((....))))))).-..)...)))).))))))....)).)). ( -19.70, z-score = -1.87, R) >droSim1.chr3R 18520324 73 - 27517382 ---GCCUAAUGGUGAAAUAUGCGGC-AGCAUAAAGCAAGAAAUUGCUUUU-AGGACAGCAUCUUCACUUUCAUAAGCA ---((.((..((((((..((((..(-.....(((((((....))))))).-..)...)))).))))))....)).)). ( -19.70, z-score = -1.87, R) >droSec1.super_0 19074269 72 - 21120651 ----CCUAAUGGUGAAAUAUGCGGC-AGCAUAAAGCAAGAAAUUGCUUUU-AGGACAGCAUCUUCACUUUCAUAAGCA ----......((((((..((((..(-.....(((((((....))))))).-..)...)))).)))))).......... ( -17.90, z-score = -1.67, R) >droYak2.chr3R 19562588 77 - 28832112 GCUUCCUAAUGGUGAAAUAUGCCGC-AGCAUAAAGCAAGAAAUUGCUUUUAAGGACAGCAUCUUCACUUUCAUAAGCA ((((......((((((..((((.((-.....(((((((....)))))))....).).)))).)))))).....)))). ( -22.10, z-score = -2.82, R) >droEre2.scaffold_4820 1109392 73 + 10470090 ---GGCUAAUGGUGAAAUAUGCCGC-AGCAUAAAGCAAGAAAUUGCUUUU-AGGACAGCAACUUCACUUUCAUAAGCA ---.(((.((((((((...(((.((-.....(((((((....))))))).-..).).)))..)))))...))).))). ( -19.70, z-score = -2.02, R) >droAna3.scaffold_13340 11084528 68 + 23697760 ---GCCUAAUGGAGAAAUAUGUGCU-GGCAUAAAGCAAGGAAUUGCUUUU-AGGACA-GCAUCU----CCUUUAAGUA ---((.(((.(((((......((((-(.(..(((((((....))))))).-..).))-))))))----)).))).)). ( -21.40, z-score = -2.44, R) >droPer1.super_145 72735 73 + 90410 ---GUCUAAUGGUGAAAUAUGCCCCUGGCAUAAAGCAAGAAAUUGCUUUU-AGAAUAG-AAGGCGGCCUCCUUAAGCA ---(..(((.((.(.....((((.(((....(((((((....))))))).-....)))-..))))..).)))))..). ( -17.90, z-score = -0.55, R) >droWil1.scaffold_181089 11044261 73 + 12369635 ---GCUUAAUGGAGAAAUAUGCCAC-AGCGUUAAGCAAGGAAUUGCUUUU-AGGACAUGCUACUGAUACUUUUAAGUA ---(((((((((.........))).-(((((.((((((....))))))..-.....)))))..........)))))). ( -11.90, z-score = 0.55, R) >consensus ___GCCUAAUGGUGAAAUAUGCCGC_AGCAUAAAGCAAGAAAUUGCUUUU_AGGACAGCAUCUUCACUUUCAUAAGCA ....(((..........(((((.....)))))((((((....))))))...)))........................ (-11.70 = -11.44 + -0.26)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:36:22 2011