| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,704,644 – 18,704,734 |
| Length | 90 |
| Max. P | 0.686907 |

| Location | 18,704,644 – 18,704,734 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 84.38 |
| Shannon entropy | 0.33466 |
| G+C content | 0.40842 |
| Mean single sequence MFE | -21.50 |
| Consensus MFE | -12.26 |
| Energy contribution | -13.47 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.686907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
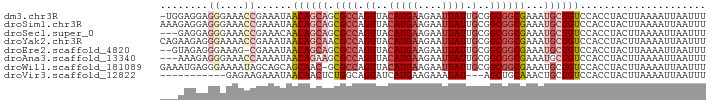
>dm3.chr3R 18704644 90 - 27905053 -UGGAGGAGGGAAACCGAAAUAACAGCAGCGCCAGUUACAUGAAGAAUUAUUGCGGCGGCGAAAUGCUGUCCACCUACUUAAAAUUAAUUU -.((.(((((....)).......(((((.((((.((..((...........))..))))))...)))))))).))................ ( -28.00, z-score = -3.44, R) >droSim1.chr3R 18519693 91 - 27517382 AAAGAGGAGGGAAACCGAAAUAACAGCAGCGCCAGUUACAUGAAGAAUUAUUGCGGCGGCGAAAUGCUGUCCACCUACUUAAAAUUAAUUU .((((((.((....))......((((((.((((.((..((...........))..))))))...))))))...))).)))........... ( -27.80, z-score = -3.81, R) >droSec1.super_0 19073652 88 - 21120651 ---GAGGAGGGAAACCGAAACAACAGCAGCGCCAGUUACAUGAAGAAUUAUUGCGGCGGCGAAAUGCUGUCCACCUACUUAAAAUUAAUUU ---.(((.((....))......((((((.((((.((..((...........))..))))))...))))))...)))............... ( -26.30, z-score = -3.37, R) >droYak2.chr3R 19561932 91 - 28832112 CAGAAGAGGGAAAACCGAAAUAACAGCAACGCCAGUUACAUGAAGAAUUAUUGCGGCGGCGAAAUGCUGUCCACCUACUUAAAAUUAAUUU ...(((.((.....))......((((((.((((.((..((...........))..))))))...)))))).......)))........... ( -18.70, z-score = -1.46, R) >droEre2.scaffold_4820 1108768 88 + 10470090 --GUAGAGGGAAAG-CGAAAUAACAGCAGCGCCAGUUACAUGAAGAAUUAUUGCGGCGGCGAAAUGCUGUCCACCUACUUAAAAUUAAUUU --((((..(((...-........(((((.((((.((..((...........))..))))))...))))))))..))))............. ( -24.20, z-score = -2.39, R) >droAna3.scaffold_13340 11083930 88 + 23697760 ---AAAGAGGGAAACCAAAAUAACAGAAGCGCCAGUUACAUGAAGAAUUAUUGCGGCGGCGAAAUGCUGUCCACCUACUUAAAAUUAAUUU ---.(((.((....))......((((...((((.((..((...........))..)))))).....)))).......)))........... ( -18.40, z-score = -1.43, R) >droWil1.scaffold_181089 11043552 90 + 12369635 GAAAUGAGGGAAAAUAGCAGCAGCAAC-GCGCCAGUUACAUGAAGAAUUAUUGCGGCGGCGAAAUGCUGUCCACCUACUUAAAAUUAAUUU ....((((........((((((.....-.((((.((..((...........))..))))))...)))))).......)))).......... ( -19.46, z-score = -0.35, R) >droVir3.scaffold_12822 3921990 77 + 4096053 -----------GAGAAGAAAUAACAACUCUGGCAGUAUCAUGAAGAAAUAU---AGCUGCAAACUGCUGUCCACCUACUUAAAAUUAAUUU -----------(((..(......)..)))((((((((...((.((......---..)).))...))))).))).................. ( -9.10, z-score = 0.08, R) >consensus ___AAGGAGGGAAACCGAAAUAACAGCAGCGCCAGUUACAUGAAGAAUUAUUGCGGCGGCGAAAUGCUGUCCACCUACUUAAAAUUAAUUU ........((....))......((((((.((((.((..((...........))..))))))...))))))..................... (-12.26 = -13.47 + 1.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:36:21 2011