| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,346,550 – 8,346,668 |
| Length | 118 |
| Max. P | 0.619624 |

| Location | 8,346,550 – 8,346,668 |
|---|---|
| Length | 118 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.25 |
| Shannon entropy | 0.56347 |
| G+C content | 0.38359 |
| Mean single sequence MFE | -28.10 |
| Consensus MFE | -10.40 |
| Energy contribution | -11.01 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.619624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8346550 118 - 23011544 -UUUUUCAUAUCAAGGCGACCGCAGUUUGCGAAUUCUGAAAUUUAACUGAAAAUCUUGUGAUUUGUUUGAGACUUUUUAUAGAGUUUUAAA-AGCGGACUGCGCUGCUACUGCGACGUCA -.............((((..((((((((((....................(((((....))))).(((((((((((....)))))))))))-.)))))))))).(((....))).)))). ( -33.80, z-score = -2.57, R) >droSec1.super_3 3842006 118 - 7220098 -UUUUUCUUAUCAAGGCGACCGCAGUUUGCGAAUUCUGAAAUUUAACUGAAAAUCUUGUGAUUUGUUUGAGACUUUUUAUAGAGUUUUAUA-AGCGGACUGCGCUGCUACUGCGACGUCA -.............((((..((((((((((....................(((((....)))))...(((((((((....)))))))))..-.)))))))))).(((....))).)))). ( -31.80, z-score = -2.09, R) >droYak2.chr2L 10984857 112 - 22324452 -------UUAUCAAGGCGACCGCAGUUUGCGAAUUCUGAAAUUUAACUGAAAAUCUUGUGAUUUGUUUGAGACUUUUUAUAGAGUUUUAAA-AGCGGACUGCGCUGCUACUGCGACGUCA -------.......((((..((((((((((....................(((((....))))).(((((((((((....)))))))))))-.)))))))))).(((....))).)))). ( -33.80, z-score = -2.92, R) >droEre2.scaffold_4929 17256177 112 - 26641161 -------UUAUCAAGGCGACCGCAGUUUGCGAAUUCUGAAAUUUAACUGAAAAUCUUGUGAUUUGUUUGAGACUUUUUAUAGAGUUUUAAA-UGCGGACUGCGCUGCUACUGCGACGUCA -------.......((((..((((((((((....................(((((....)))))((((((((((((....)))))))))))-))))))))))).(((....))).)))). ( -34.90, z-score = -3.48, R) >droAna3.scaffold_12916 10263018 100 + 16180835 -----UUUAUUUUAGUCGACCGCUGGUUGCAAAUUCAGAAAUUUAACUGAAAAUCUUGUGAUUUGUUAGAGACUUUUUAUAUGG---UGGGCUGCGACUGCGACGUCA------------ -----............((((((.(((((((..(((((........))))).(((.(((((...(((...)))...))))).))---)....))))))))))..))).------------ ( -22.10, z-score = -0.58, R) >dp4.chr4_group3 7935929 107 - 11692001 -------GUGCUGGGUCGACCGCAGUUUGCAAAUUCUCAAAUUUAACUGAAAAUCUUGUGAUUUGUUUCAGACUUUUUAUAGAGUUUUAUACUGCGACAGCGGCAACGGCGCCA------ -------((((((.((((..((((((.((.(((((((.........(((((((((....)))...)))))).........))))))))).))))))....))))..))))))..------ ( -32.17, z-score = -2.41, R) >droPer1.super_1 9383243 107 - 10282868 -------GUGCUGGGUCGACCGCAGUUUGCAAAUUCUCAAAUUUAACUGAAAAUCUUGUGAUUUGUUUCAGACUUUUUAUAGAGUUUUAUACUGCGACAGCGGCUACGGCGCCA------ -------(((((((((((..((((((.((.(((((((.........(((((((((....)))...)))))).........))))))))).))))))....))))).))))))..------ ( -33.67, z-score = -2.98, R) >droWil1.scaffold_180703 2421571 101 + 3946847 UUUUUGGUUUCUUAGUUGACCGUCGAUUGCAAAUUCUGAAAUUUAACUGAAAAUCUUGUGAUUUGUUUGAGACUUUUUAUAGAGUUGUGGCUACCGCACAU------------------- .....(((((.((((((((...((((........)).))...))))))))))))).((((....(((...((((((....))))))..))).....)))).------------------- ( -18.80, z-score = -0.42, R) >droVir3.scaffold_12963 16113567 102 - 20206255 ---CUUCACUU----GCGACCGCAGUUUACGAAUUCUGGAAUUUAACUGAAAAUCUUGUGAUUUGUUUGAGACUUUUUAUAGAGUUUUAUA-AGCGACUUGCGCAGCGAC---------- ---..((.(((----((((.(((.....(((((((..(((.(((.....))).)))...))))))).(((((((((....)))))))))..-.)))..))))).)).)).---------- ( -20.70, z-score = -0.40, R) >droMoj3.scaffold_6500 5575443 103 - 32352404 ---CUUCAGCU-UACGCGACCGCAGUUUAUGAAUUCUGAAAUUUAACUGAAAAUCUUGUGAUUUGUUUGAGACUUUUUAUAGAGUUUUAUA-AGCCAGUUGCGCAGCG------------ ---.....(((-..((((((....(((((((((((((((((.........(((((....)))))...........))).))))).))))))-)))..)))))).))).------------ ( -22.25, z-score = -0.78, R) >droGri2.scaffold_15252 13543594 116 - 17193109 ---UUUCAGUUUUUUGCGACCGCUUUUUGCGAAUUCUGGAAUUUAACUGAAAAUCUUCUGAUUUGUUUGAGACAUUUUAUAGAGUUUUAUA-AGCGAGUUUCGCAGCGACUGCGACACAA ---((((((((..((.(((.(((.....)))...)).).))...)))))))).......((((((((((.((.((((....)))).)).))-))))))))((((((...))))))..... ( -25.10, z-score = -0.42, R) >consensus ____UUCUUAUUAAGGCGACCGCAGUUUGCGAAUUCUGAAAUUUAACUGAAAAUCUUGUGAUUUGUUUGAGACUUUUUAUAGAGUUUUAUA_AGCGAACUGCGCUGCGACUGCG______ ....................(((.....(((((((.(((.((((......)))).))).))))))).(((((((((....)))))))))....)))........................ (-10.40 = -11.01 + 0.61)

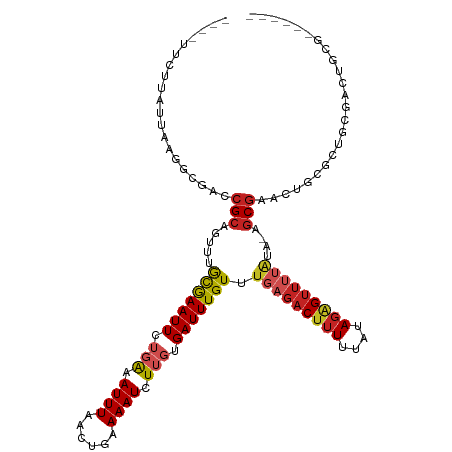

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:25:45 2011