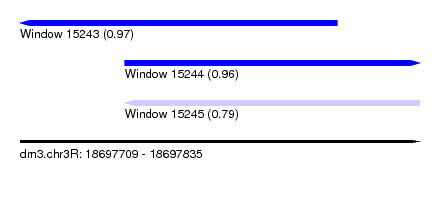
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,697,709 – 18,697,835 |
| Length | 126 |
| Max. P | 0.965527 |

| Location | 18,697,709 – 18,697,809 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 76.76 |
| Shannon entropy | 0.42645 |
| G+C content | 0.29798 |
| Mean single sequence MFE | -17.94 |
| Consensus MFE | -8.43 |
| Energy contribution | -8.57 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.965527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18697709 100 - 27905053 UUUCUGUAAAUUAACAUAGAAAUAUGUAAUUUCCGCUAUGGGCAGGUAUAAAAAAAAAAAUAAAUGCAAAAAGAAAGAAAAGCGAAAGGUAUAAGCGGAA-- .............(((((....)))))...(((((((....(((..(((..........)))..)))...............(....).....)))))))-- ( -19.20, z-score = -3.14, R) >droEre2.scaffold_4820 1101765 92 + 10470090 UUUCUGUAAAUUAACAUAGAAAUAUGUAAUUUCCGCUAUGGGCAGGU-------AAAAAAAAGAUGU-AAAGGAAAUAGAAGCGAAAAGUAUAAGCGGAA-- .((((((......(((((....))))).((((((.......(((...-------..........)))-...)))))).................))))))-- ( -14.62, z-score = -1.48, R) >droSec1.super_0 19059431 96 - 21120651 UUUCUGUAAAUUAACAUAGAAAUAUGUAAUUUCCGCUAUGGGCAGGUAUA----AAAAAAUAAAUGCAAAAAGAAAGAAAGCCGAAAGGUAUAAGCGGAA-- .............(((((....)))))...(((((((....(((..(((.----.....)))..))).............(((....)))...)))))))-- ( -25.10, z-score = -4.95, R) >droSim1.chr3R 18512879 96 - 27517382 UUUCUGUAAAUUAACAUAGAAAUAUGUAAUUUCCGCUAUGGGCAGGUAUA----AAAAAAUAAAUGCAAAAAGAAAGAAAAGCGAAAGGUAUAAGCGGAA-- .............(((((....)))))...(((((((....(((..(((.----.....)))..)))...............(....).....)))))))-- ( -20.30, z-score = -3.40, R) >droAna3.scaffold_13340 11077522 75 + 23697760 UUUCUGUAAAUUAACAUAGAAAUAUGUAAUUUCCGUUAUGG---------------------AAUG----AAAAAGAAAAGGCGAAAAGUAUAAGCGGAA-- .((((((......(((((....)))))...((((.....))---------------------))..----........................))))))-- ( -12.00, z-score = -1.91, R) >droWil1.scaffold_181089 11036150 94 + 12369635 UUUCAGCAAAUUAACAUAGAAAUAUGUAAUUUCCGUUCUGUUCUGG--------GAGGGAAAACAGCAAAAAAAAAGGAAGAAAAAAAGAAAAUAGGGCCCA .....((......(((((....)))))..(((((.((((.....))--------)).)))))...))................................... ( -16.40, z-score = -1.08, R) >consensus UUUCUGUAAAUUAACAUAGAAAUAUGUAAUUUCCGCUAUGGGCAGGU_______AAAAAAUAAAUGCAAAAAGAAAGAAAAGCGAAAAGUAUAAGCGGAA__ .............(((((....)))))....((((((........................................................))))))... ( -8.43 = -8.57 + 0.14)

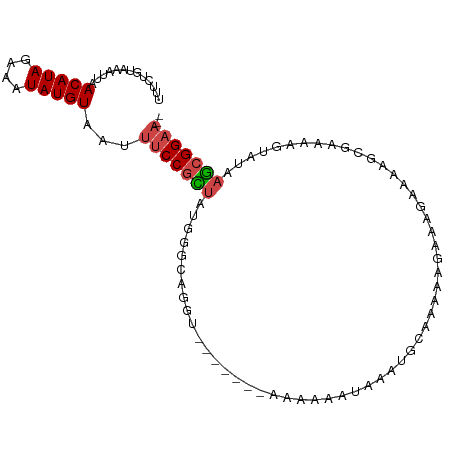
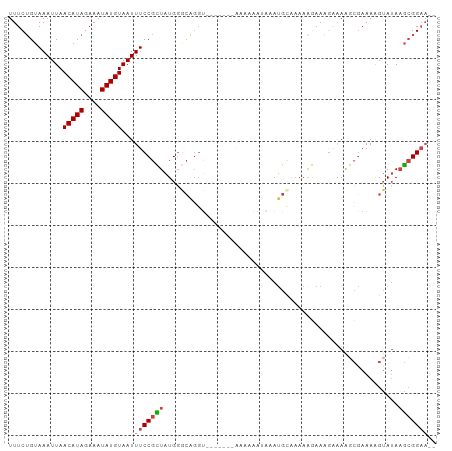
| Location | 18,697,742 – 18,697,835 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 79.67 |
| Shannon entropy | 0.35590 |
| G+C content | 0.36767 |
| Mean single sequence MFE | -21.90 |
| Consensus MFE | -13.82 |
| Energy contribution | -15.74 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.961194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18697742 93 + 27905053 GCAUUUAUUUUUUUUUUUAUACCUGCCCAUAGCGGAAAUUACAUAUUUCUAUGUUAAUUUACAGAAAUAGAG----GUGCAGGUGUAGAAGGCAACG ..............((((((((((((((.....((((((.....)))))).(((......)))........)----).))))))))))))(....). ( -24.80, z-score = -3.70, R) >droEre2.scaffold_4820 1101797 86 - 10470090 ------ACAUCUUUUUUU-UACCUGCCCAUAGCGGAAAUUACAUAUUUCUAUGUUAAUUUACAGAAAUAGAG----GUGCAGGUGUGGAAGGCAGCG ------...((((((...-(((((((((.....((((((.....)))))).(((......)))........)----).))))))).))))))..... ( -21.50, z-score = -1.89, R) >droSec1.super_0 19059464 89 + 21120651 ----GCAUUUAUUUUUUUAUACCUGCCCAUAGCGGAAAUUACAUAUUUCUAUGUUAAUUUACAGAAAUAGAG----GUGCAGGUGUGGAAGGCAGCG ----........((((((((((((((((.....((((((.....)))))).(((......)))........)----).))))))))))))))..... ( -24.00, z-score = -2.59, R) >droSim1.chr3R 18512912 89 + 27517382 ----GCAUUUAUUUUUUUAUACCUGCCCAUAGCGGAAAUUACAUAUUUCUAUGUUAAUUUACAGAAAUAGAG----GUGCAGGUGUGGAAGGCAGCG ----........((((((((((((((((.....((((((.....)))))).(((......)))........)----).))))))))))))))..... ( -24.00, z-score = -2.59, R) >droWil1.scaffold_181089 11036185 79 - 12369635 --------GCUGUUUUCCCUCCCAGAACAGAACGGAAAUUACAUAUUUCUAUGUUAAUUUGCUGAAAUAGAAAAUAGUGCAGGUGCC---------- --------(((((((((.....(((.....(((((((((.....))))))..)))......))).....)))))))))((....)).---------- ( -15.20, z-score = -1.07, R) >consensus ______AUUUAUUUUUUUAUACCUGCCCAUAGCGGAAAUUACAUAUUUCUAUGUUAAUUUACAGAAAUAGAG____GUGCAGGUGUGGAAGGCAGCG .............(((((((((((((.(.....((((((.....)))))).(((......))).............).)))))))))))))...... (-13.82 = -15.74 + 1.92)
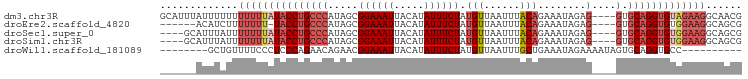
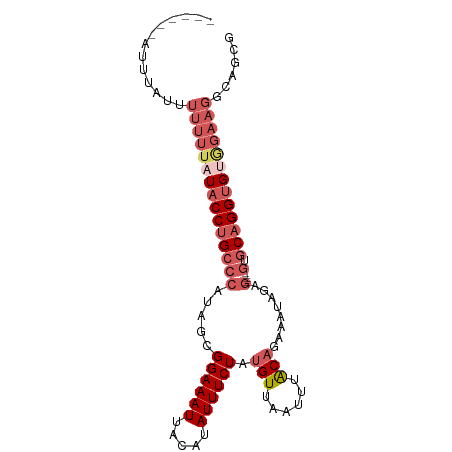
| Location | 18,697,742 – 18,697,835 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 79.67 |
| Shannon entropy | 0.35590 |
| G+C content | 0.36767 |
| Mean single sequence MFE | -17.20 |
| Consensus MFE | -11.78 |
| Energy contribution | -12.58 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.791213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18697742 93 - 27905053 CGUUGCCUUCUACACCUGCAC----CUCUAUUUCUGUAAAUUAACAUAGAAAUAUGUAAUUUCCGCUAUGGGCAGGUAUAAAAAAAAAAAUAAAUGC .............((((((.(----(........((.((((((.((((....)))))))))).))....)))))))).................... ( -17.50, z-score = -2.05, R) >droEre2.scaffold_4820 1101797 86 + 10470090 CGCUGCCUUCCACACCUGCAC----CUCUAUUUCUGUAAAUUAACAUAGAAAUAUGUAAUUUCCGCUAUGGGCAGGUA-AAAAAAAGAUGU------ .............((((((.(----(........((.((((((.((((....)))))))))).))....)))))))).-............------ ( -17.50, z-score = -1.57, R) >droSec1.super_0 19059464 89 - 21120651 CGCUGCCUUCCACACCUGCAC----CUCUAUUUCUGUAAAUUAACAUAGAAAUAUGUAAUUUCCGCUAUGGGCAGGUAUAAAAAAAUAAAUGC---- .............((((((.(----(........((.((((((.((((....)))))))))).))....))))))))................---- ( -17.50, z-score = -1.78, R) >droSim1.chr3R 18512912 89 - 27517382 CGCUGCCUUCCACACCUGCAC----CUCUAUUUCUGUAAAUUAACAUAGAAAUAUGUAAUUUCCGCUAUGGGCAGGUAUAAAAAAAUAAAUGC---- .............((((((.(----(........((.((((((.((((....)))))))))).))....))))))))................---- ( -17.50, z-score = -1.78, R) >droWil1.scaffold_181089 11036185 79 + 12369635 ----------GGCACCUGCACUAUUUUCUAUUUCAGCAAAUUAACAUAGAAAUAUGUAAUUUCCGUUCUGUUCUGGGAGGGAAAACAGC-------- ----------.((....))................((......(((((....)))))..(((((.((((.....)))).)))))...))-------- ( -16.00, z-score = -1.10, R) >consensus CGCUGCCUUCCACACCUGCAC____CUCUAUUUCUGUAAAUUAACAUAGAAAUAUGUAAUUUCCGCUAUGGGCAGGUAUAAAAAAAAAAAU______ .............((((((..................((((((.((((....))))))))))((.....)))))))).................... (-11.78 = -12.58 + 0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:36:19 2011