| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,676,180 – 18,676,281 |
| Length | 101 |
| Max. P | 0.974320 |

| Location | 18,676,180 – 18,676,281 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 60.75 |
| Shannon entropy | 0.76442 |
| G+C content | 0.49372 |
| Mean single sequence MFE | -27.63 |
| Consensus MFE | -10.50 |
| Energy contribution | -11.71 |
| Covariance contribution | 1.21 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.974320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
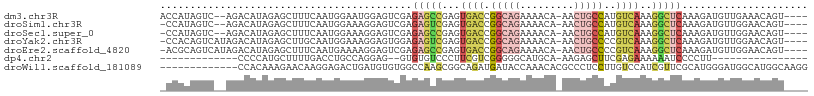
>dm3.chr3R 18676180 101 - 27905053 ACCAUAGUC--AGACAUAGAGCUUUCAAUGGAAUGGAGUCGAGAGCCGAGUGACCGGCAGAAAACA-AACUGCCAUGUCAAAGGCUCAAAGAUGUUGAAACAGU---- ......((.--.(((((...(.(((((......))))).)..(((((...((((.(((((......-..)))))..))))..)))))....)))))...))...---- ( -28.60, z-score = -2.40, R) >droSim1.chr3R 18490938 100 - 27517382 -CCAUAGUC--AGACAUAGAGCUUUCAAUGGAAAGGAGUCGAGAGUCGAGUGACCGGCAGAAAACA-AACUGCCAUGUCAAAGGCUCAAAGAUGUUGGAACAGU---- -(((..(((--.(((......(((((....)))))..)))..(((((...((((.(((((......-..)))))..))))..)))))...)))..)))......---- ( -30.00, z-score = -2.92, R) >droSec1.super_0 19037605 100 - 21120651 -CCAUAGUC--AGACAUAGAGCUUUCAAUGGAAAGGAGUCGAGAGCCGAGUGACCGGCAGAAAACA-AACUGCCAUGUCAAAGGCUCAAAGAUGUUGGAACAGU---- -(((..(((--.(((......(((((....)))))..)))..(((((...((((.(((((......-..)))))..))))..)))))...)))..)))......---- ( -32.70, z-score = -3.77, R) >droYak2.chr3R 19531327 102 - 28832112 -CCACAGUCAUAGACAUAGAGCUUGCAAUGGAAAGGAGUGGAGAGUCGAGUGACCGGCAGAAAACA-AACUGCCCCGUCAAAGGCUCAAAGAUGUUGGAACAGU---- -((((.(((...)))......(((........)))..)))).(((((...((((.(((((......-..)))))..))))..))))).................---- ( -27.30, z-score = -1.43, R) >droEre2.scaffold_4820 1079910 102 + 10470090 -ACGCAGUCAUAGACAUAGAGCUUUCAAUGAAAAGGAGUCGAGAGCCGAGUGACCGGCAGAAAACA-AACUGCCCCGUCAAAGGCUCAAAGAUGUUGGAACAGU---- -......((...(((((.((.((((.......))))..))..(((((...((((.(((((......-..)))))..))))..)))))....))))).)).....---- ( -27.90, z-score = -1.98, R) >dp4.chr2 24666734 76 + 30794189 -------------CCCCAUGCUUUUGACCUGCCAGGAG--GUGUGUCCCUUCGUCGGGGGCAUGCA-AAGAGCUUCGAGAAAAAAUCCCCUU---------------- -------------......((((((..((.....))..--((((((((((.....)))))))))).-))))))...................---------------- ( -21.00, z-score = 0.23, R) >droWil1.scaffold_181089 11018881 95 + 12369635 -------------CCACAAAGAACAAGGAGACUGAUGUGUGGCCAAGCGGCAGAUGAUACCAAACACGCCCUCCUUGUCCAUCGUUCGCAUGGGAUGGCAUGGCAAGG -------------(((......((((((((......((((((((....)))...((....))..))))).))))))))(((((.(......).)))))..)))..... ( -25.90, z-score = 0.47, R) >consensus _CCAUAGUC__AGACAUAGAGCUUUCAAUGGAAAGGAGUCGAGAGCCGAGUGACCGGCAGAAAACA_AACUGCCAUGUCAAAGGCUCAAAGAUGUUGGAACAGU____ ..........................................(((((...((((.(((((.........)))))..))))..)))))..................... (-10.50 = -11.71 + 1.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:36:14 2011