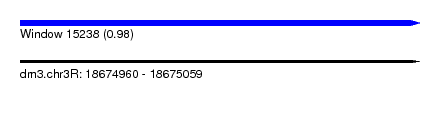
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,674,960 – 18,675,059 |
| Length | 99 |
| Max. P | 0.980208 |

| Location | 18,674,960 – 18,675,059 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 68.73 |
| Shannon entropy | 0.57957 |
| G+C content | 0.55356 |
| Mean single sequence MFE | -28.06 |
| Consensus MFE | -17.91 |
| Energy contribution | -17.40 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18674960 99 + 27905053 GUGAGUGGUCCACAUUCCUGGC------UGUGGAAUGUGAUCCCCGCAAUUUUCGGCCAUUUCCAUUAC---GGCCCUGCACUCCAACCAACACC--CACCGACUUUCCC--------- (.((((((..((((((((....------...))))))))..))..(((......((((...........---)))).))))))))..........--.............--------- ( -24.90, z-score = -1.01, R) >droPer1.super_145 43865 93 - 90410 GUGGGGGGUCUGCAUUCCUGCUGCCUCAUGUGGAAUGCGAUACCCACAAUUUUCGGCCAUUUCCAUUAUGUGGGCCCC-GAGGGCAGCCUCUUC------------------------- (((((..(((.(((((((.((........)))))))))))).))))).......(((.....((((...))))((((.-..)))).))).....------------------------- ( -36.10, z-score = -1.32, R) >dp4.chr2 24665809 94 - 30794189 GUGGGGGGUCUGCAUUCCUGCUGCCUCAUGUGGAAUGCGAUACCCACAAUUUUGGGCCAUUUCCAUUAUUGGGGCCCCCGGGGGCAGCCUCUUC------------------------- ..((((((((.(((((((.((........)))))))))))).(((.((((..((((.....))))..))))))))))))((((....))))...------------------------- ( -40.50, z-score = -1.33, R) >droAna3.scaffold_13340 11056355 91 - 23697760 CUGGGCGGUCGACAUUCCUGCC------CGUGGAAUGUGAUCUCCGCAAUUUUCGGCCAUUUCCAUUAC---GACUACCCCGACUCCCUUUUUCC--UACUU----------------- ..(((..(((((((((((....------...))))))).....(((.......)))............)---)))..)))...............--.....----------------- ( -22.50, z-score = -1.68, R) >droEre2.scaffold_4820 1078647 99 - 10470090 GUGAGUGGUCCGCAUUCCUGGC------UGUGGAAUGUGAUCCCCGCAAUUUUCGGCCAUUUCCAUUAC---GGCCCUGCACUCCAAACUCCACC--CACCGACUUUUCC--------- (((.((((..((((((((....------...))))))))......(((......((((...........---)))).)))..........)))).--)))..........--------- ( -26.70, z-score = -1.56, R) >droYak2.chr3R 19530071 101 + 28832112 AUGAGUGGUCCACAUUCCUGGC------CGUGGAAUGUGAUCCCCGCAAUUUUCGGCCAUUUCCAUUAC---GGCCCUGCACUCCAACUCCAAGCACCACAGAAUUUUCG--------- ....(((((.((((((((....------...))))))))......(((......((((...........---)))).)))...............)))))..........--------- ( -25.60, z-score = -1.17, R) >droSec1.super_0 19036376 99 + 21120651 GUGAGUGGUCCACAUUCCUGGC------UGUGGAAUGUGAUCCCCGCAAUUUUCGGCCAUUUCCAUUAU---GGCCCUGCACUCCAAACCCCACA--UACCGACUUUUCC--------- (((.((((..((((((((....------...))))))))......(((......((((((.......))---)))).)))..........)))).--)))..........--------- ( -28.60, z-score = -2.38, R) >droSim1.chr3R 18489715 99 + 27517382 GUGAGUGGUCCACAUUCCUGGC------UGUGGAAUGUGAUCCCCGCAAUUUUCGGCCAUUUCCAUUAC---GGCCCUGCACUCCAAACCCCGCC--CACCGACUUUUCC--------- (((.((((..((((((((....------...))))))))......(((......((((...........---)))).)))..........)))).--)))..........--------- ( -26.90, z-score = -1.58, R) >droWil1.scaffold_181089 11017530 103 - 12369635 -----UGGUCGGCAUUCCUGCC------CGUGGAAUGUUCGUUCCGCAAUUUUGGGCCAUUUCCAUUAC---GGUUCCCUUAUCCCCCUUCCACC--CCCCCACCUCACGCUCUCUCUC -----(((((((((....))))------.(((((((....))))))).......)))))..........---.......................--...................... ( -20.70, z-score = -0.70, R) >consensus GUGAGUGGUCCACAUUCCUGGC______UGUGGAAUGUGAUCCCCGCAAUUUUCGGCCAUUUCCAUUAC___GGCCCUGCACUCCAACCUCCACC__CACCGACUUUUCC_________ ....((((..((((((((.............))))))))....)))).......((((..............))))........................................... (-17.91 = -17.40 + -0.50)

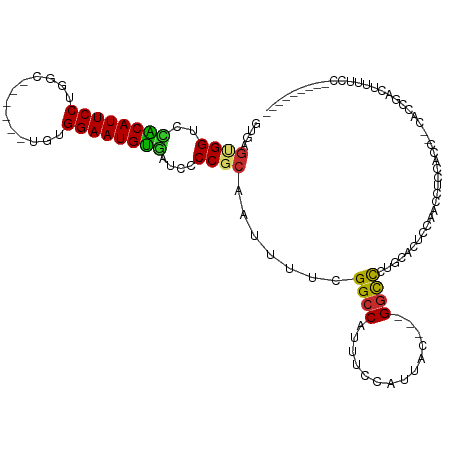
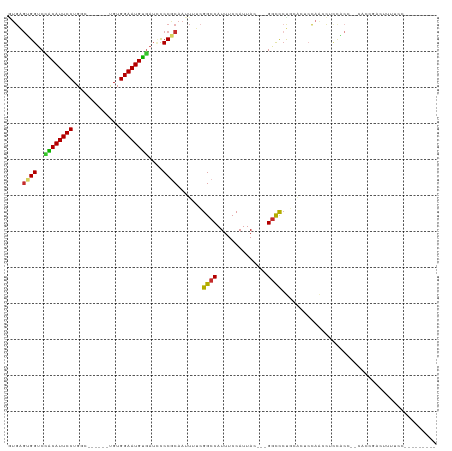
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:36:13 2011