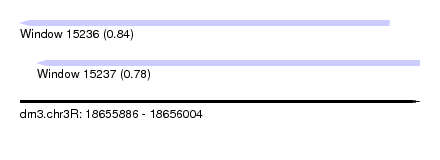
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,655,886 – 18,656,004 |
| Length | 118 |
| Max. P | 0.835175 |

| Location | 18,655,886 – 18,655,995 |
|---|---|
| Length | 109 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 85.62 |
| Shannon entropy | 0.31148 |
| G+C content | 0.44706 |
| Mean single sequence MFE | -28.32 |
| Consensus MFE | -19.82 |
| Energy contribution | -19.60 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.835175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18655886 109 - 27905053 CGAUAAAAA-CCAAAGGAGAACUUCGCACAGGAAGUCGCUAU-GGGGCGAAAAUCUGAUGCGGAAAUCGAAAUUUGUUAAAUGGAGCAAUCGAUGGAUGGC---GCUGACUGAC-- .........-(((.........((((((((((...(((((..-..)))))...)))).)))))).(((((...(((((......))))))))))...))).---..........-- ( -26.40, z-score = -1.42, R) >droPer1.super_145 16268 103 + 90410 CAAUAAAAGCUUAAGGAAAAACUUCGCACCGGAAGUCGCUAUGGGGGCGAAAAUCUGAUGCGGAAAUCGAAAUUUGUUAAAUGGAGCAAUCGAUGGAUGGGGC------------- ........((((..........((((((.((((..(((((.....)))))...)))).)))))).(((((...(((((......)))))))))).....))))------------- ( -26.70, z-score = -2.19, R) >droSim1.chr3R 18469428 109 - 27517382 CGAUAAAAA-CCAAAGGAGAACUUCGCACAGGAAGUCGCUAU-GGGGCGAAAAUCUGAUGCGGAAAUCGAAAUUUGUUAAAUGGAGCAAUCGAUGGAUGGC---GAUGACUGAC-- .........-(((.........((((((((((...(((((..-..)))))...)))).)))))).(((((...(((((......))))))))))...))).---..........-- ( -26.40, z-score = -1.79, R) >droSec1.super_0 19017421 109 - 21120651 CGAUAAAAA-CCAAAGGAGAACUUCGCACAGGAAGUCGCUAU-GGGGCGAAAAUCUGAUGCGGAAAUCGAAAUUUGUUAAAUGGAGCAAUCGAUGGAUGGC---GAUGACUGAC-- .........-(((.........((((((((((...(((((..-..)))))...)))).)))))).(((((...(((((......))))))))))...))).---..........-- ( -26.40, z-score = -1.79, R) >droYak2.chr3R 19510060 109 - 28832112 CGAUAAAAA-CCGAAGGAGAACUUCGCACAGGAAGUCGCUAU-GGGGCGAAAAUCUGAUGCGGAAAUCGAAAUUUGUUAAAUGGAGCAAUCGAUGGAUGGC---GAUGACUGAC-- .........-((((((.....))))(((((((...(((((..-..)))))...)))).)))))..((((..(((..((.....((....))))..)))..)---))).......-- ( -28.90, z-score = -2.43, R) >droEre2.scaffold_4820 1059088 109 + 10470090 CGAUAAAAA-CCAAAGCAGAACUUCGCACAGGAAGUCGCUAU-GGGGCGAAAAUCUGAUGCGGAAAUCGAAAUUUGUUAAAUGGAGCAAUCGAUGGAUGGC---GAUGACUGAC-- .........-......(((...((((((((((...(((((..-..)))))...)))).)))))).((((..(((..((.....((....))))..)))..)---)))..)))..-- ( -28.50, z-score = -2.22, R) >droAna3.scaffold_13340 11038263 112 + 23697760 CGAUAAAAA-CCAAAGGAAAACUUCGCACAGGAAGUCGCUAU-GGGGCGAAAAUCUGAUGCGGAAAUCGAAAUUUGUUAAAUGGAGCAAUCGAUGGAUGGCGCUGCUGACUGAC-- .........-(((.........((((((((((...(((((..-..)))))...)))).)))))).(((((...(((((......))))))))))...)))..............-- ( -26.40, z-score = -1.14, R) >droWil1.scaffold_181089 10995523 109 + 12369635 CAAUAAAAAGCCAAAGAAGAAGUUCGCACAGGAAGUCGCUAU-GGGGCGAAAAUCUGAUGCGGAAAUCGAAAUUUGUUAAAUGGAGCAAUCGAUGGAUGGC---UCUGGCACA--- .........(((......((..((((((((((...(((((..-..)))))...)))).))))))..))..............(((((.(((....))).))---))))))...--- ( -35.10, z-score = -3.84, R) >droMoj3.scaffold_6540 1177327 113 + 34148556 CAAUAAAAGCGUUAGG-AAAACUUCGCGCAGGAAAUCGCUAU-GGGGCGAAAAUCUGUAGCGGAAAUCGAAAUUUGUUAAAUGGAGCAAUCGAUGAAUUGCG-UUGGCUCUGACAG ..........((((((-..(((((((((((((...(((((..-..)))))...))))).))))).(((((...(((((......)))))))))).......)-))..).))))).. ( -31.00, z-score = -1.87, R) >droVir3.scaffold_12822 3886015 112 + 4096053 CAAUAAAAGCAUGAAC-AAAACUUUGCACAGGAAGUCGCUAU-GGGGCGAAAAUCUGAAGCGGAAAUCGAAAUUUGUUAAAUGGAGCAAUCGAUGAAUAGCG-UUGGAUCUGACG- ........(((.....-.......))).((((...(((((..-..)))))...((..(.((....(((((...(((((......)))))))))).....)).-)..))))))...- ( -23.50, z-score = -0.70, R) >droGri2.scaffold_15074 3459729 114 - 7742996 CAAUAAAAGCGGAGGCAAAAACUUUCCACAGGAAGUUGCCAU-GGGGCGAAAAUCUGAAGCGGAAAUCGAAAUUUGUUAAAUGGAGCAAUCGAUGAAUCGCGCUUGGCCCUGGUG- .........(((.(((............((((...(((((..-..)))))...))))(((((((.(((((...(((((......))))))))))...)).))))).))))))...- ( -32.20, z-score = -1.24, R) >consensus CGAUAAAAA_CCAAAGGAGAACUUCGCACAGGAAGUCGCUAU_GGGGCGAAAAUCUGAUGCGGAAAUCGAAAUUUGUUAAAUGGAGCAAUCGAUGGAUGGC___GAUGACUGAC__ ......................(((((...(((..(((((.....)))))...)))...))))).(((((...(((((......))))))))))...................... (-19.82 = -19.60 + -0.22)

| Location | 18,655,891 – 18,656,004 |
|---|---|
| Length | 113 |
| Sequences | 11 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 87.29 |
| Shannon entropy | 0.27737 |
| G+C content | 0.44525 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -21.60 |
| Energy contribution | -21.47 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.784430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18655891 113 - 27905053 UAGGAGGAGCGAUAAAAA-CCAAAGGAGAACUUCGCACAGGAAGUCGCUAU-GGGGCGAAAAUCUGAUGCGGAAAUCGAAAUUUGUUAAAUGGAGCAAUCGAUGGAUGGCGCUGA--- .......((((.......-((..........((((((((((...(((((..-..)))))...)))).)))))).(((((...(((((......))))))))))))....))))..--- ( -30.00, z-score = -1.94, R) >droPer1.super_145 16269 111 + 90410 GGGGGAAAGCAAUAAAAGCUUAAGGAAAAACUUCGCACCGGAAGUCGCUAUGGGGGCGAAAAUCUGAUGCGGAAAUCGAAAUUUGUUAAAUGGAGCAAUCGAUGGAUGGGG------- ......((((.......))))..........((((((.((((..(((((.....)))))...)))).)))))).(((((...(((((......))))))))))........------- ( -27.00, z-score = -1.81, R) >droSim1.chr3R 18469433 113 - 27517382 UAGGAGGAGCGAUAAAAA-CCAAAGGAGAACUUCGCACAGGAAGUCGCUAU-GGGGCGAAAAUCUGAUGCGGAAAUCGAAAUUUGUUAAAUGGAGCAAUCGAUGGAUGGCGAUGA--- ........((.((.....-............((((((((((...(((((..-..)))))...)))).)))))).(((((...(((((......))))))))))..)).)).....--- ( -27.50, z-score = -1.66, R) >droSec1.super_0 19017426 113 - 21120651 UAGGAGGAGCGAUAAAAA-CCAAAGGAGAACUUCGCACAGGAAGUCGCUAU-GGGGCGAAAAUCUGAUGCGGAAAUCGAAAUUUGUUAAAUGGAGCAAUCGAUGGAUGGCGAUGA--- ........((.((.....-............((((((((((...(((((..-..)))))...)))).)))))).(((((...(((((......))))))))))..)).)).....--- ( -27.50, z-score = -1.66, R) >droYak2.chr3R 19510065 113 - 28832112 UAGGAGGAGCGAUAAAAA-CCGAAGGAGAACUUCGCACAGGAAGUCGCUAU-GGGGCGAAAAUCUGAUGCGGAAAUCGAAAUUUGUUAAAUGGAGCAAUCGAUGGAUGGCGAUGA--- ........((.((.....-((((((.....))))(((((((...(((((..-..)))))...)))).)))))..(((((...(((((......))))))))))..)).)).....--- ( -30.40, z-score = -2.41, R) >droEre2.scaffold_4820 1059093 113 + 10470090 UGGGAGGAGCGAUAAAAA-CCAAAGCAGAACUUCGCACAGGAAGUCGCUAU-GGGGCGAAAAUCUGAUGCGGAAAUCGAAAUUUGUUAAAUGGAGCAAUCGAUGGAUGGCGAUGA--- (((...............-)))..((.....((((((((((...(((((..-..)))))...)))).)))))).(((((...(((((......)))))))))).....)).....--- ( -27.66, z-score = -1.15, R) >droAna3.scaffold_13340 11038268 116 + 23697760 UAGGAGAAGCGAUAAAAA-CCAAAGGAAAACUUCGCACAGGAAGUCGCUAU-GGGGCGAAAAUCUGAUGCGGAAAUCGAAAUUUGUUAAAUGGAGCAAUCGAUGGAUGGCGCUGCUGA ....((.((((.......-((..........((((((((((...(((((..-..)))))...)))).)))))).(((((...(((((......))))))))))))....)))).)).. ( -31.00, z-score = -1.97, R) >droWil1.scaffold_181089 10995527 114 + 12369635 UAGGAAAAGCAAUAAAAAGCCAAAGAAGAAGUUCGCACAGGAAGUCGCUAU-GGGGCGAAAAUCUGAUGCGGAAAUCGAAAUUUGUUAAAUGGAGCAAUCGAUGGAUGGCUCUGG--- ........((........))((((...((..((((((((((...(((((..-..)))))...)))).))))))..))....))))......(((((.(((....))).)))))..--- ( -31.40, z-score = -2.87, R) >droMoj3.scaffold_6540 1177336 113 + 34148556 UGGGAUAAGCAAUAAAAGCGUUAGG-AAAACUUCGCGCAGGAAAUCGCUAU-GGGGCGAAAAUCUGUAGCGGAAAUCGAAAUUUGUUAAAUGGAGCAAUCGAUGAAUUGCGUUGG--- ........(((((....((((.(((-....))).))))......(((((((-(((.......))))))))))..(((((...(((((......))))))))))..))))).....--- ( -28.50, z-score = -1.68, R) >droVir3.scaffold_12822 3886023 110 + 4096053 ---GAUAAGCAAUAAAAGCAUGAAC-AAAACUUUGCACAGGAAGUCGCUAU-GGGGCGAAAAUCUGAAGCGGAAAUCGAAAUUUGUUAAAUGGAGCAAUCGAUGAAUAGCGUUGG--- ---......((((....(((.....-.......))).((((...(((((..-..)))))...))))..((....(((((...(((((......)))))))))).....)))))).--- ( -22.80, z-score = -0.90, R) >droGri2.scaffold_15074 3459737 115 - 7742996 UGGGAUCAGCAAUAAAAGCGGAGGCAAAAACUUUCCACAGGAAGUUGCCAU-GGGGCGAAAAUCUGAAGCGGAAAUCGAAAUUUGUUAAAUGGAGCAAUCGAUGAAUCGCGCUUGG-- ...............(((((((((.......))))).((((...(((((..-..)))))...))))..((((..(((((...(((((......))))))))))...))))))))..-- ( -29.20, z-score = -0.83, R) >consensus UAGGAGAAGCGAUAAAAA_CCAAAGGAGAACUUCGCACAGGAAGUCGCUAU_GGGGCGAAAAUCUGAUGCGGAAAUCGAAAUUUGUUAAAUGGAGCAAUCGAUGGAUGGCGAUGA___ ........((.....................(((((...(((..(((((.....)))))...)))...))))).(((((...(((((......)))))))))).....))........ (-21.60 = -21.47 + -0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:36:13 2011