
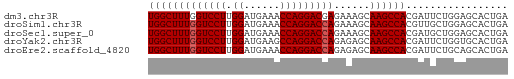
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,648,516 – 18,648,576 |
| Length | 60 |
| Max. P | 0.842100 |

| Location | 18,648,516 – 18,648,576 |
|---|---|
| Length | 60 |
| Sequences | 5 |
| Columns | 60 |
| Reading direction | forward |
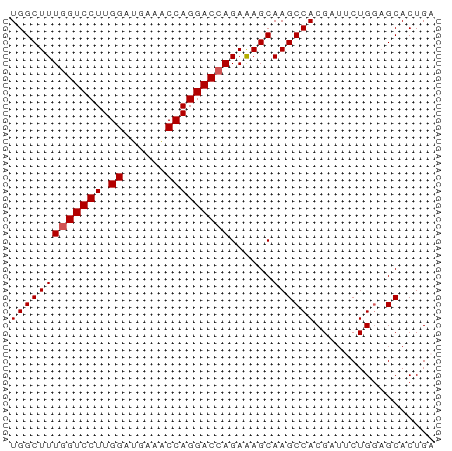
| Mean pairwise identity | 94.67 |
| Shannon entropy | 0.09253 |
| G+C content | 0.53333 |
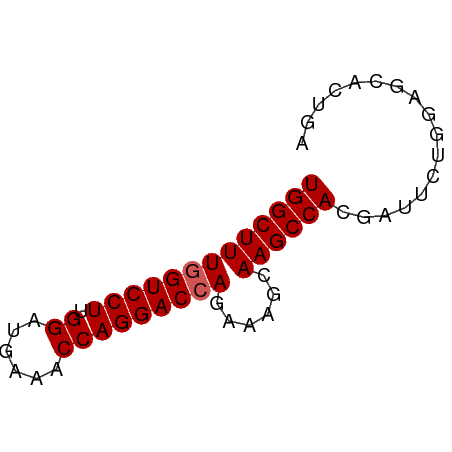
| Mean single sequence MFE | -20.28 |
| Consensus MFE | -17.86 |
| Energy contribution | -18.06 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.744093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
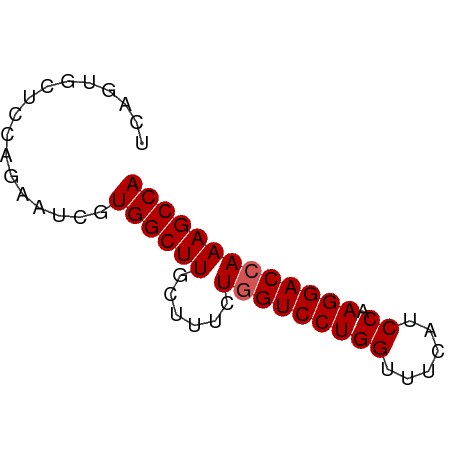
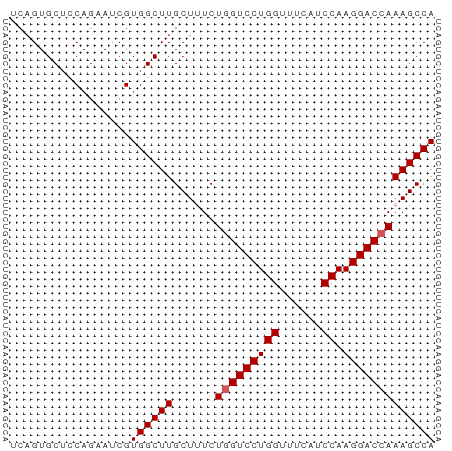
>dm3.chr3R 18648516 60 + 27905053 UGGCUUUGGUCCUUGGAUGAAACCAGGACGAGAAAGCAAGCCACGAUUCUGGAGCACUGA ..(((((.(((((.((......)))))))...)))))..((..((....))..))..... ( -15.20, z-score = -0.44, R) >droSim1.chr3R 18460781 60 + 27517382 UGGCUUUGGUCCUUGGAUGAAACCAGGACCAGAAAGCAAGCCACGUUGCUGGAGCACUGA (((((((((((((.((......)))))))))......))))))((.(((....))).)). ( -22.00, z-score = -2.15, R) >droSec1.super_0 19009291 60 + 21120651 UGGCUUUGGUCCUUGGAUGAAACCAGGACCAGAAAGCAAGCCACGAUGCUGGAGCACUGA (((((((((((((.((......)))))))))......))))))((.(((....))).)). ( -22.00, z-score = -2.65, R) >droYak2.chr3R 19501519 60 + 28832112 UGGCUUUGGUCCUUGGAUGAAGCCAGGACCAGAGAGCAAGCCACGAUUCUGGUGCACUGA ...((((((((((.((......)))))))))))).((..((((......))))))..... ( -22.90, z-score = -2.05, R) >droEre2.scaffold_4820 1046574 60 - 10470090 UGGCUUUGGUCCUUGGAUGAAACCAGGACCAGAGAGCAAGCCACGAUUCUGCAGCACUGA ...((((((((((.((......)))))))))))).(((...........)))........ ( -19.30, z-score = -1.48, R) >consensus UGGCUUUGGUCCUUGGAUGAAACCAGGACCAGAAAGCAAGCCACGAUUCUGGAGCACUGA (((((((((((((.((......)))))))))......))))))................. (-17.86 = -18.06 + 0.20)
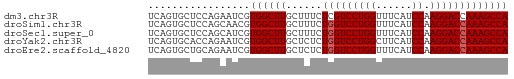
| Location | 18,648,516 – 18,648,576 |
|---|---|
| Length | 60 |
| Sequences | 5 |
| Columns | 60 |
| Reading direction | reverse |
| Mean pairwise identity | 94.67 |
| Shannon entropy | 0.09253 |
| G+C content | 0.53333 |
| Mean single sequence MFE | -19.34 |
| Consensus MFE | -17.72 |
| Energy contribution | -17.92 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.842100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18648516 60 - 27905053 UCAGUGCUCCAGAAUCGUGGCUUGCUUUCUCGUCCUGGUUUCAUCCAAGGACCAAAGCCA .....((..(......)..))..(((((...(((((((......)).))))).))))).. ( -14.70, z-score = -0.81, R) >droSim1.chr3R 18460781 60 - 27517382 UCAGUGCUCCAGCAACGUGGCUUGCUUUCUGGUCCUGGUUUCAUCCAAGGACCAAAGCCA ....(((....)))...((((((......(((((((((......)).))))))))))))) ( -21.30, z-score = -2.28, R) >droSec1.super_0 19009291 60 - 21120651 UCAGUGCUCCAGCAUCGUGGCUUGCUUUCUGGUCCUGGUUUCAUCCAAGGACCAAAGCCA ...((((....))))..((((((......(((((((((......)).))))))))))))) ( -22.10, z-score = -2.77, R) >droYak2.chr3R 19501519 60 - 28832112 UCAGUGCACCAGAAUCGUGGCUUGCUCUCUGGUCCUGGCUUCAUCCAAGGACCAAAGCCA .................((((((......(((((((((......)).))))))))))))) ( -18.20, z-score = -0.95, R) >droEre2.scaffold_4820 1046574 60 + 10470090 UCAGUGCUGCAGAAUCGUGGCUUGCUCUCUGGUCCUGGUUUCAUCCAAGGACCAAAGCCA .....((..(......)..))..(((...(((((((((......)).))))))).))).. ( -20.40, z-score = -1.68, R) >consensus UCAGUGCUCCAGAAUCGUGGCUUGCUUUCUGGUCCUGGUUUCAUCCAAGGACCAAAGCCA .................((((((......(((((((((......)).))))))))))))) (-17.72 = -17.92 + 0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:36:09 2011