
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,645,461 – 18,645,554 |
| Length | 93 |
| Max. P | 0.679088 |

| Location | 18,645,461 – 18,645,554 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 64.49 |
| Shannon entropy | 0.67607 |
| G+C content | 0.51973 |
| Mean single sequence MFE | -17.58 |
| Consensus MFE | -6.73 |
| Energy contribution | -7.43 |
| Covariance contribution | 0.71 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.679088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
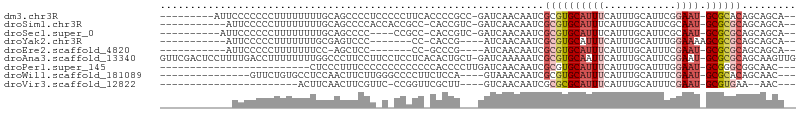
>dm3.chr3R 18645461 93 + 27905053 ---------AUUCCCCCCCUUUUUUUUGCAGCCCCUCCCCCUUCACCCCGCC-GAUCAACAAUCGCGUGCAUUUCAUUUGCAUUCGGAAU-GCGCACAGCAGCA-- ---------.................(((.((.................((.-(((.....)))))(((((((((..........)))))-))))...)).)))-- ( -17.20, z-score = -1.84, R) >droSim1.chr3R 18457694 90 + 27517382 -----------AUUCCCCCUUUUUUUUGCAGCCCCACCACCGCC-CACCGUC-GAUCAACAAUCGCGUGCAUUUCAUUUGCAUUCGCAAU-GCGCGCAGCAGCA-- -----------...............(((.((............-.......-(((.....)))((((((((......(((....)))))-)))))).)).)))-- ( -18.30, z-score = -1.52, R) >droSec1.super_0 19006229 87 + 21120651 ----------AUUCCCCCCUUUUUUUUGCAGCCCC----CCGCC-CACCGUC-GAUCAACAAUCGCGUGCAUUUCAUUUGCAUUCGCAAU-GCGCGCAGCAGCA-- ----------................(((.((...----.....-.......-(((.....)))((((((((......(((....)))))-)))))).)).)))-- ( -18.30, z-score = -1.60, R) >droYak2.chr3R 19498448 81 + 28832112 -----------AUUCCCCCUUUUUUUGCGAGUCCC-------CC-CACCG----AUCAACAAUCGCGUGCAUUUCAUUUGCAUUUGGAAAAGCGCGCAGCAGCA-- -----------.............((((.......-------..-....(----((.....)))((((((.(((((........)))))..)))))).))))..-- ( -16.90, z-score = -0.90, R) >droEre2.scaffold_4820 1043597 79 - 10470090 -----------AUUCCCCCUUUUUUUCC-AGCUCC-------CC-GCCCG----AUCAACAAUCGCGUGCAUUUCAUUUGCAUUUCGAAU-GCGCGCAGCAGCA-- -----------.................-.(((..-------..-((..(----((.....)))((((((((((.(........).))))-)))))).))))).-- ( -17.50, z-score = -2.35, R) >droAna3.scaffold_13340 11027831 104 - 23697760 GUUCGACUCCUUUUGACCUUUUUUUUGGCCCUUCCUUCCUCCUCACACUGCU-GAUCAAAAAUCGCGUGCAAUUCAUUUGCAUUCGGAAU-GCGCGCAGCAAGUUG ...(((((........((........))....................((((-(((.....)))(((((((.(((..........))).)-))))))))))))))) ( -20.00, z-score = -0.46, R) >droPer1.super_145 4005 77 - 90410 -------------------------CUCCCUUUCCCCCCCCCCCCCACCCCUUGAUCAACAAUCGCGUGCAUUUCAUUUGCAUUUGGAAU-GCGGGCGGCAAC--- -------------------------.....................................((((.(((((((((........))))))-))).))))....--- ( -17.90, z-score = -1.19, R) >droWil1.scaffold_181089 10984703 83 - 12369635 ---------------GUUCUGUGCCUCCAACUUCUUGGGCCCCUUCUCCA----GUAAACAAUCGCGUGCAUUUCAUUUGCAUUUCGAAU-GCGCACAGCAAC--- ---------------(((((((((.(((((....)))))...........----.....((.(((.(((((.......)))))..))).)-).)))))).)))--- ( -19.90, z-score = -1.92, R) >droVir3.scaffold_12822 3877469 72 - 4096053 -----------------------ACUUCAACUUCGUUC-CCGGUUCGCUU----GUCAACAAUCGCGCGCAUUUCAUUUGCAUUUCGAAU-GCGUGAA--AAC--- -----------------------..((((....(((((-...((.((.((----(....))).)).))(((.......))).....))))-)..))))--...--- ( -12.20, z-score = 0.44, R) >consensus ____________UUCCCCCUUUUUUUUCCAGUUCCU_C_CCCCC_CACCG___GAUCAACAAUCGCGUGCAUUUCAUUUGCAUUUGGAAU_GCGCGCAGCAGCA__ ................................................................((((((((((............)))).))))))......... ( -6.73 = -7.43 + 0.71)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:36:08 2011