
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,643,810 – 18,643,941 |
| Length | 131 |
| Max. P | 0.746492 |

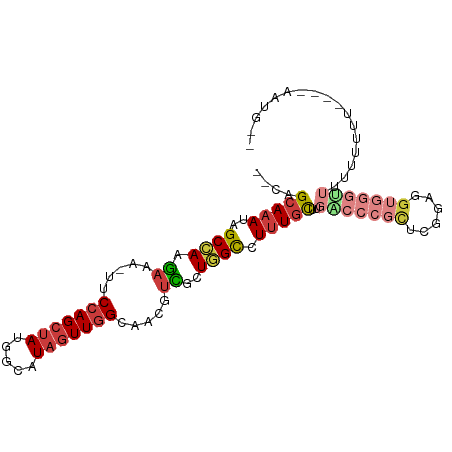
| Location | 18,643,810 – 18,643,905 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 70.83 |
| Shannon entropy | 0.56387 |
| G+C content | 0.46520 |
| Mean single sequence MFE | -27.49 |
| Consensus MFE | -16.80 |
| Energy contribution | -18.50 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.32 |
| Mean z-score | -0.79 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.746492 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 18643810 95 + 27905053 --CAGCAAAUAGCCAAGAAA-UUCCAGCUAUGGCAUAGUUGGCAACGUCGCUGGCCUUUGCUUGACCCGCUCGGAGGUGGGUUUUUUUUUUUUUAAUG-- --.((((((..((((.((..-((((((((((...)))))))).))..))..)))).)))))).((((((((....))))))))...............-- ( -31.30, z-score = -2.06, R) >droAna3.scaffold_13340 11026597 94 - 23697760 UUUUGCAAACAGCCAAGAAA-UUCCAGCUAUUCCAUAGUUGGCAACGUUGGUCGCCUUUGUUUUGCC--UCUGGACGUUUGCUUGGAGAGGGAAAGA--- ....((((((.(((((....-..((((((((...)))))))).....))))).)..)))))(((.((--(((...((......)).))))).)))..--- ( -21.60, z-score = 1.03, R) >droEre2.scaffold_4820 1042072 88 - 10470090 --CAGCAAACAGCCAAGAAAAUGCCAGCUAUGGCAUAGUUGGCAGUGGCGCUGGCCUUUGCUUGGCCCGCUUGGAGGUGGGUUUUAAUGG---------- --.((((((..((((.(....((((((((((...))))))))))....)..)))).)))))).((((((((....)))))))).......---------- ( -35.60, z-score = -1.21, R) >droYak2.chr3R 19496830 92 + 28832112 --CAGCAAAUAGCCAAGAAAAUUCCAGCUAUGGCAUAGUUGGCAACGUCGCUGGCCUUUGCUUGACCCGCUUGGAGGUGGUGGCUUUUU-----AAUGG- --((..(((.(((((.......((.(((...(((.((((((....))..)))))))...))).)).(((((....)))))))))).)))-----..)).- ( -28.10, z-score = -0.15, R) >droSec1.super_0 19004629 93 + 21120651 --CAGCAAAUAGCCAAGAAA-UUCCAGCUAUGGCAUAGUUGGCAACGUCGCUGGCCUUUGCUUGACCCGCUCGGAGGUGGGUUUUUUUUU----AAUGUG --.((((((..((((.((..-((((((((((...)))))))).))..))..)))).)))))).((((((((....)))))))).......----...... ( -31.30, z-score = -1.80, R) >droSim1.chr3R 18456082 93 + 27517382 --CAGCAAAUAGCCAAGAAA-UUCCAGCUAUGGCAUAGUUGGCAACGUCGCUGGCCUUUGCUUGACCCGCUCGGAGGUGGGUUUUUUUUUU---AAUGU- --.((((((..((((.((..-((((((((((...)))))))).))..))..)))).)))))).((((((((....))))))))........---.....- ( -31.30, z-score = -1.84, R) >droGri2.scaffold_15074 3448303 75 + 7742996 --AAGAAAAUGGCAAAAA-----CCAGAAACGCCAUAGUUUGCUUUUUUUUUUGUGUGUGUGUG--------GGGGGUUUUUUUUUUUUU---------- --(((((((..((.....-----(((...(((((((((.............))))).)))).))--------)...))..)))))))...---------- ( -13.22, z-score = 0.50, R) >consensus __CAGCAAAUAGCCAAGAAA_UUCCAGCUAUGGCAUAGUUGGCAACGUCGCUGGCCUUUGCUUGACCCGCUCGGAGGUGGGUUUUUUUUU____AAUG__ ....(((((..((((.((.....(((((((.....))))))).....))..)))).)))))..(((((((......)))))))................. (-16.80 = -18.50 + 1.70)


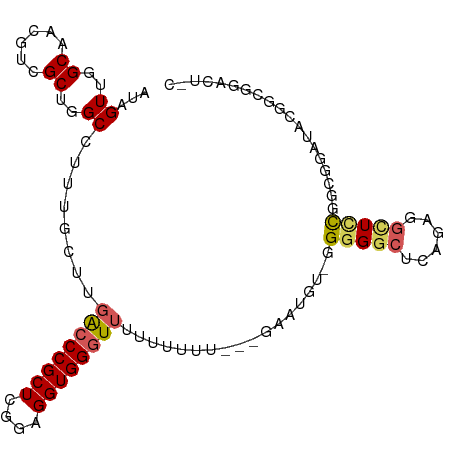
| Location | 18,643,841 – 18,643,941 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 72.60 |
| Shannon entropy | 0.48746 |
| G+C content | 0.57702 |
| Mean single sequence MFE | -35.82 |
| Consensus MFE | -18.60 |
| Energy contribution | -19.04 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.623098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18643841 100 + 27905053 AUAGUUGGCAACGUCGCUGGCCUUUGCUUGACCCGCUCGGAGGUGGGUUUUUUUUUUUUUAAUG--GGGGACUCAGAGGCUUCGGCGGAUACGGCGGACUCC ..((((.((..(((((..((((((((...((((((((....))))))))..............(--(....)))))))))).)))))......)).)))).. ( -37.40, z-score = -1.92, R) >droEre2.scaffold_4820 1042104 78 - 10470090 AUAGUUGGCAGUGGCGCUGGCCUUUGCUUGGCCCGCUUGGAGGUGGGUUUUAAUGGG--GGCUCCUGGAGG---AGGAGCUCC------------------- ......(((((.(((....))).))))).((((((((....))))))))......((--(((((((.....---)))))))))------------------- ( -35.30, z-score = -1.66, R) >droYak2.chr3R 19496862 90 + 28832112 AUAGUUGGCAACGUCGCUGGCCUUUGCUUGACCCGCUUGGAGGUGG-UGGCUUUUUAAUGGGUGG-UGGGGCUCAGUGGUUCUAGGGGCUAC---------- .....((((..(.(((((((((.(..((..(((((...((((((..-..))))))...)))))))-..)))).)))))).....)..)))).---------- ( -30.20, z-score = -0.22, R) >droSec1.super_0 19004660 98 + 21120651 AUAGUUGGCAACGUCGCUGGCCUUUGCUUGACCCGCUCGGAGGUGGGUUUUUUUUU----AAUGUGGGGGGCUCAGAGGCUCCGGCGAAUACGGCGGACUCC ..((((.((....(((((((((((..(..((((((((....)))))))).......----...)..))))(((....))).))))))).....)).)))).. ( -37.60, z-score = -1.07, R) >droSim1.chr3R 18456113 98 + 27517382 AUAGUUGGCAACGUCGCUGGCCUUUGCUUGACCCGCUCGGAGGUGGGUUUUUUUUU---UAAUGU-GGGGGCUCAGAGGCUCCGGCGGAUACGGCGGACUAC ......(....)(((.((((((((..(..((((((((....)))))))).......---....).-.))))).))).)))((((.((....)).)))).... ( -38.62, z-score = -2.00, R) >consensus AUAGUUGGCAACGUCGCUGGCCUUUGCUUGACCCGCUCGGAGGUGGGUUUUUUUUU___GAAUGU_GGGGGCUCAGAGGCUCCGGCGGAUACGGCGGACU_C ...((..((......))..))........((((((((....))))))))..................(((((......)))))................... (-18.60 = -19.04 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:36:07 2011