
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,643,229 – 18,643,305 |
| Length | 76 |
| Max. P | 0.821785 |

| Location | 18,643,229 – 18,643,305 |
|---|---|
| Length | 76 |
| Sequences | 10 |
| Columns | 79 |
| Reading direction | forward |
| Mean pairwise identity | 92.82 |
| Shannon entropy | 0.15060 |
| G+C content | 0.37951 |
| Mean single sequence MFE | -20.09 |
| Consensus MFE | -15.17 |
| Energy contribution | -15.14 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.821785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18643229 76 + 27905053 GAAAUAACAGCGCAAAGUACAAGUAAAGUGUAUUUUCA--CAUUGAAUGAGGUUAAGCUUCGUUAAAUUUU-GCGCAGC .........(((((((((.........(((......))--)....(((((((.....)))))))..)))))-))))... ( -21.20, z-score = -3.30, R) >droAna3.scaffold_13340 11026024 76 - 23697760 GAAAUGACAGCGCAAAGUAGAGGUAAAGUGUAUUUUCA--CAUUGAAUGAGGUUAAGCUUCGUUAAAUUUU-CCGCAGC .........(((.(((((.........(((......))--)....(((((((.....)))))))..)))))-.)))... ( -14.60, z-score = -0.65, R) >droEre2.scaffold_4820 1041507 76 - 10470090 GAAAUGACAGCGCAAAGUACAAGUAAAGUGUAUUUUCA--CAUUGAAUGAGGUUAAGCUUCGUUAAAUUUU-GCGCAGC .........(((((((((.........(((......))--)....(((((((.....)))))))..)))))-))))... ( -21.20, z-score = -2.83, R) >droYak2.chr3R 19496229 76 + 28832112 GAAAUGACAGCGCAAAGUACAAGUAAAGUGUAUUUUCA--CAUUGAAUGAGGUUAAGCUUCGUUAAAUUUU-GCGCAGC .........(((((((((.........(((......))--)....(((((((.....)))))))..)))))-))))... ( -21.20, z-score = -2.83, R) >droSec1.super_0 19004023 76 + 21120651 GAAAUGACAGCGCAAAGUACAAGUAAAGUGUAUUUUCA--CAUUGAAUGAGGUUAAGCUUCGUUAAAUUUU-GCGCAGC .........(((((((((.........(((......))--)....(((((((.....)))))))..)))))-))))... ( -21.20, z-score = -2.83, R) >droSim1.chr3R 18455479 76 + 27517382 GAAAUGACAGCGCAAAGUACAAGUAAAGUGUAUUUUCA--CAUUGAAUGAGGUUAAGCUUCGUUAAAUUUU-GCGCAGC .........(((((((((.........(((......))--)....(((((((.....)))))))..)))))-))))... ( -21.20, z-score = -2.83, R) >droWil1.scaffold_181089 10982610 77 - 12369635 GAAAUGACAGCGCAAAGUACACGUAAAGUGUAUUUUCA--CAUUGAAUGAGGUUAAGCUUCGUUAAAUUUUUGCGCAGC .........(((((((((((((.....)))))).....--.....(((((((.....))))))).....)))))))... ( -22.60, z-score = -2.93, R) >droVir3.scaffold_12822 3875482 78 - 4096053 GAAAUGACAGUCCAAAGUGUAAGUAAAGUGGAUUUUUUGCCAUUGAGCGAGGUUAAGCUUCGUUAAAUUUU-GCGCAGC ................(((((((...(((((........))))).(((((((.....)))))))....)))-))))... ( -17.90, z-score = -1.12, R) >droMoj3.scaffold_6540 1161665 77 - 34148556 GAAAUGACAGCGCAAAGUGUACGUAAAGUGGAUUUUUU-CCAUUGAACGAGGUUAAGCUUCGUUAAAUUUC-GCACAGC ................((((......((((((.....)-))))).(((((((.....))))))).......-))))... ( -18.20, z-score = -1.29, R) >droGri2.scaffold_15074 3447243 76 + 7742996 GAAAUGACAGCGCAAAGUGUACGUAAAGUGUAUUUUCC--CAUUGAACGAGGUUAAGCUUCGUUAAAUUUU-GCGCAGC .........((((((((((((((.....))))).....--.....(((((((.....)))))))..)))))-))))... ( -21.60, z-score = -2.54, R) >consensus GAAAUGACAGCGCAAAGUACAAGUAAAGUGUAUUUUCA__CAUUGAAUGAGGUUAAGCUUCGUUAAAUUUU_GCGCAGC .........((((((((((((.......)))))))).........(((((((.....)))))))........))))... (-15.17 = -15.14 + -0.03)

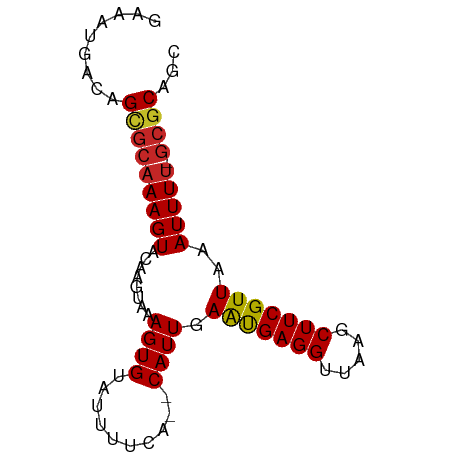
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:36:05 2011